
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,296,404 – 13,296,560 |
| Length | 156 |
| Max. P | 0.909475 |

| Location | 13,296,404 – 13,296,520 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13296404 116 - 22224390 CGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGCUAGAACAACUAUAGCAGCGAAUAU----AUCACAUUGAUAUAAUAGUAGUAAUUAAUGAAACUUUAG ...(((((.....)))))(((((((.((((((((((....))))))((..(((((.........)))))))...(((----(((.....))))))....))))...)))))))....... ( -24.10) >DroSec_CAF1 11655 112 - 1 CGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGUUAGAACAAUUAUAGCAGCGUAUAU----AUCACAUUGAUAUAAUA-UAGUAAUGAAUGAAAC---AG .(((((((.....))))(((((((((....((((((....))))))((..(((((.........)))))))...(((----(((.....))))))...-..)))))))))..)))---.. ( -24.50) >DroSim_CAF1 5562 112 - 1 CGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGUUAAAACAACUAUAGCAGCGUAUAU----AUCACAUUGAUAUAAUA-UAGUAAUGAAUGAAAC---AG .(((((((.....))))(((((((((....((((((....))))))((..(((((.........)))))))...(((----(((.....))))))...-..)))))))))..)))---.. ( -23.70) >DroEre_CAF1 6568 91 - 1 CGUUAUCGUAACACGGUAUUCAUUACGCUAACGAUGUUUGCAUUGU-------UACAAUAAUUAUAGAAGUG-AUA------UAAUAUUGU---A---------AUUAAUGAGAC---AG (((((.(((((...........))))).))))).(((((.((((((-------(((((((.(((((......-.))------)))))))))---)---------).)))))))))---). ( -22.10) >DroYak_CAF1 5815 103 - 1 CGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGU-------UCGAAUAAUUCUACAAGUGCAUAUAUCUAUCACAUUGAUUUAACA-C----AUUUAUGUGA-----A ...(((((.....)))))..............(((((.((((((..-------..((.....))....)))))).)))))..((((((.(((......-.----))).))))))-----. ( -19.30) >consensus CGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUG_UAGAACAAUUAUAGCAGCGUAUAU____AUCACAUUGAUAUAAUA_UAGUAAUUAAUGAAAC___AG ...(((((.....)))))((((((......((((((....))))))...................................(((.....)))...............))))))....... ( -8.93 = -9.54 + 0.61)

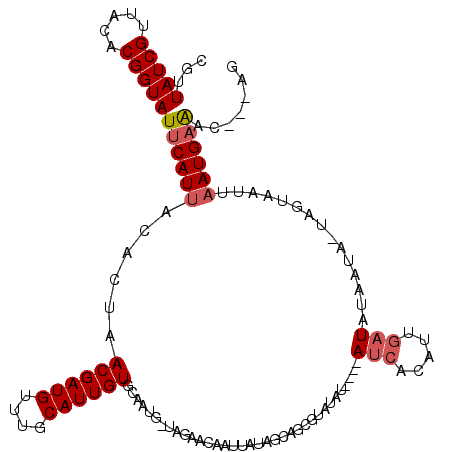
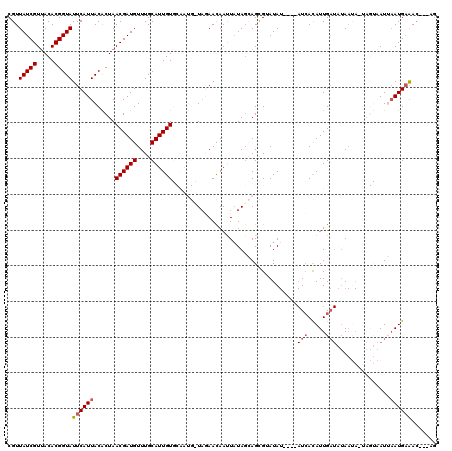
| Location | 13,296,443 – 13,296,560 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13296443 117 + 22224390 ---AUAUUCGCUGCUAUAGUUGUUCUAGCAUUGCACAAUGCAAACAUCGUUAGUGUAAUGAAUACCGUGUAACGAUAACGCAUUAUCGUAACCAAUGUGACCAUGUUUGCAAACGAAGCA ---...((((((((((.........)))))..))....(((((((((.((((((((.....)))).(.((.(((((((....))))))).)))....)))).)))))))))...)))... ( -28.20) >DroSec_CAF1 11690 117 + 1 ---AUAUACGCUGCUAUAAUUGUUCUAACAUUGCACAAUGCAAACAUCGUUAGUGUAAUGAAUACCGUGUAACGAUAACGAUUUAUCGUAACCAAUGUGACCAUGUUUGCAAACGAAGCA ---......(((......(((((.(.......).)))))((((((((.((((((((.....)))).(.((.(((((((....))))))).)))....)))).))))))))......))). ( -26.30) >DroSim_CAF1 5597 117 + 1 ---AUAUACGCUGCUAUAGUUGUUUUAACAUUGCACAAUGCAAACAUCGUUAGUGUAAUGAAUACCGUGUAACGAUAACGCUUUAUCGUUACCAAUGUGACCAUGUUUGCAAACGAAGCA ---......(((((.((.((((...)))))).))....(((((((((.((((((((.....)))).(.((((((((((....)))))))))))....)))).))))))))).....))). ( -30.70) >DroEre_CAF1 6591 108 + 1 ----UAU-CACUUCUAUAAUUAUUGUA-------ACAAUGCAAACAUCGUUAGCGUAAUGAAUACCGUGUUACGAUAACGCUUUAUCGUAACCAAUGUGACCAUGUUUGCAAACGAAGCA ----...-..((((((((.....))))-------....(((((((((.((((.(((.(((.....)))((((((((((....))))))))))..))))))).)))))))))...)))).. ( -27.20) >DroYak_CAF1 5845 113 + 1 GAUAUAUGCACUUGUAGAAUUAUUCGA-------ACAAUGCAAACAUCGUUAGUGUAAUGAAUACCGUGUAACGAUAACGCUUUAUCGUAACCAAUGUGACCAUGUUCGCAAGCGAAGCA ......((((.((((.((.....))..-------))))))))....(((((.(((..(((..(((...((.(((((((....))))))).))....)))..)))...))).))))).... ( -23.40) >consensus ___AUAUACGCUGCUAUAAUUGUUCUA_CAUUGCACAAUGCAAACAUCGUUAGUGUAAUGAAUACCGUGUAACGAUAACGCUUUAUCGUAACCAAUGUGACCAUGUUUGCAAACGAAGCA ......................................(((((((((.((((((((.....)))).(.((.(((((((....))))))).)))....)))).)))))))))......... (-20.44 = -20.84 + 0.40)


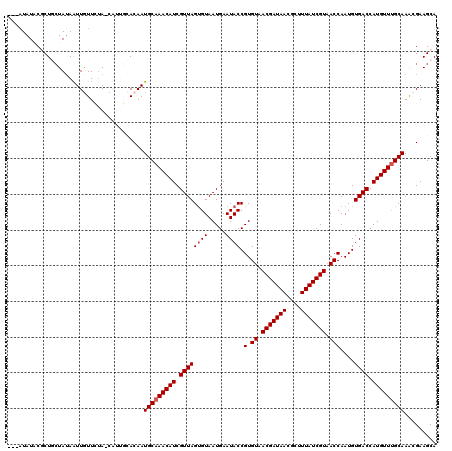
| Location | 13,296,443 – 13,296,560 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.09 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -22.84 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13296443 117 - 22224390 UGCUUCGUUUGCAAACAUGGUCACAUUGGUUACGAUAAUGCGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGCUAGAACAACUAUAGCAGCGAAUAU--- ...(((((.(((((((((.((.......((.(((((((....))))))).))...(((.....)))....)).)))))))))........(((((.........))))))))))...--- ( -29.60) >DroSec_CAF1 11690 117 - 1 UGCUUCGUUUGCAAACAUGGUCACAUUGGUUACGAUAAAUCGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGUUAGAACAAUUAUAGCAGCGUAUAU--- .........(((((((((.((.......((.(((((((....))))))).))...(((.....)))....)).))))))))).(((((..(((((.........)))))..))))).--- ( -27.70) >DroSim_CAF1 5597 117 - 1 UGCUUCGUUUGCAAACAUGGUCACAUUGGUAACGAUAAAGCGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUGUUAAAACAACUAUAGCAGCGUAUAU--- .........(((((((((.((.......((((((((((....))))))))))...(((.....)))....)).))))))))).(((((..(((((.........)))))..))))).--- ( -30.90) >DroEre_CAF1 6591 108 - 1 UGCUUCGUUUGCAAACAUGGUCACAUUGGUUACGAUAAAGCGUUAUCGUAACACGGUAUUCAUUACGCUAACGAUGUUUGCAUUGU-------UACAAUAAUUAUAGAAGUG-AUA---- .(((((...(((((((((.((.......((((((((((....))))))))))...(((.....)))....)).)))))))))((((-------....)))).....))))).-...---- ( -29.40) >DroYak_CAF1 5845 113 - 1 UGCUUCGCUUGCGAACAUGGUCACAUUGGUUACGAUAAAGCGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGU-------UCGAAUAAUUCUACAAGUGCAUAUAUC .....((((((((((((..((.((((((((.(((((((....))))))).))...(((.....))).....))))))..))..)))-------)))..........))))))........ ( -24.70) >consensus UGCUUCGUUUGCAAACAUGGUCACAUUGGUUACGAUAAAGCGUUAUCGUUACACGGUAUUCAUUACACUAACGAUGUUUGCAUUGUGCAAUG_UAGAACAAUUAUAGCAGCGUAUAU___ .(((.(...(((((((((.((.......((.(((((((....))))))).))...(((.....)))....)).)))))))))((((...........)))).....).)))......... (-22.84 = -22.40 + -0.44)

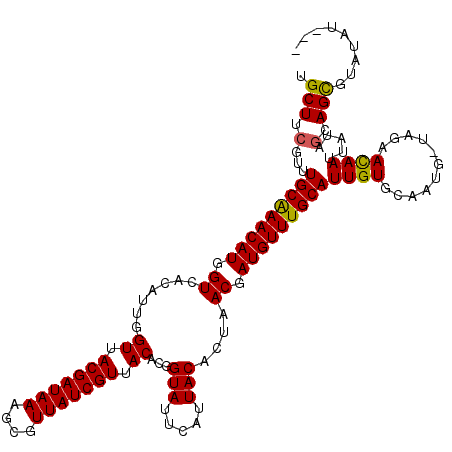
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:48 2006