
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,283,168 – 13,283,297 |
| Length | 129 |
| Max. P | 0.999618 |

| Location | 13,283,168 – 13,283,258 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13283168 90 + 22224390 GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUG ((((.(((.....((((....))))..........(((((.(((((....))))).....)))))..........)))))))........ ( -23.10) >DroSec_CAF1 29352 90 + 1 GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCUGAUAAAGUACUGCUUUCACUUUUUG ((((.(((.....((((....))))........((.((((.(((((....))))).....)))))).........)))))))........ ( -22.40) >DroSim_CAF1 30057 90 + 1 GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCGAGAUAAAGUACUGCUUUCACUUUUUG ((((((......((((((....(((....))).)))))))))))).....((((((.((.(((.((........))))).)).)))))). ( -20.60) >DroEre_CAF1 29326 90 + 1 GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUG ((((.(((.....((((....))))..........(((((.(((((....))))).....)))))..........)))))))........ ( -23.10) >DroYak_CAF1 29046 90 + 1 GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAUGAAAGAGAUGGCCAGAUAAAGUACUUCUUUCACUUUUUG (((.(((......((((....))))..........((((((((((..........))))))))))......))).)))............ ( -23.40) >consensus GAAAUGCAAAAUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUG ((((((......((((((....(((....))).)))))))))))).....((((((.((.(((.((........))))).)).)))))). (-17.76 = -18.36 + 0.60)



| Location | 13,283,168 – 13,283,258 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13283168 90 - 22224390 CAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC ...((((.((..((((........)).))..)).))))...((..(((...((((((.(((....)))....)))))).)))..)).... ( -22.40) >DroSec_CAF1 29352 90 - 1 CAAAAAGUGAAAGCAGUACUUUAUCAGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC ......(..(((((............((((((....((((....))))))))))(((.(((....)))....))).)).)))..)..... ( -21.90) >DroSim_CAF1 30057 90 - 1 CAAAAAGUGAAAGCAGUACUUUAUCUCGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC ...((((.((..((((........)).))..)).))))...((..(((...((((((.(((....)))....)))))).)))..)).... ( -22.40) >DroEre_CAF1 29326 90 - 1 CAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC ...((((.((..((((........)).))..)).))))...((..(((...((((((.(((....)))....)))))).)))..)).... ( -22.40) >DroYak_CAF1 29046 90 - 1 CAAAAAGUGAAAGAAGUACUUUAUCUGGCCAUCUCUUUCAUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC .....(((((((((..(.((......))..)..)))))))))(..(((...((((((.(((....)))....)))))).)))..)..... ( -24.30) >consensus CAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAUUUUGCAUUUC ...((((.((..((((........)).))..)).))))...((..(((...((((((.(((....)))....)))))).)))..)).... (-19.82 = -20.22 + 0.40)

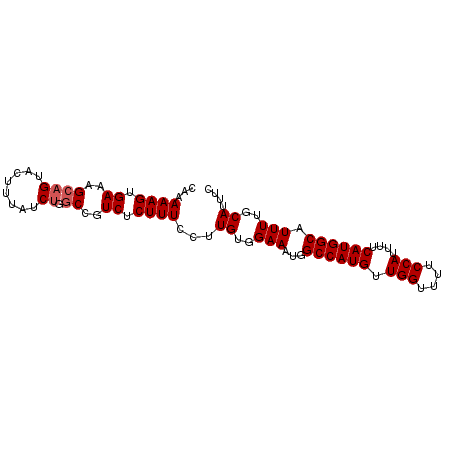
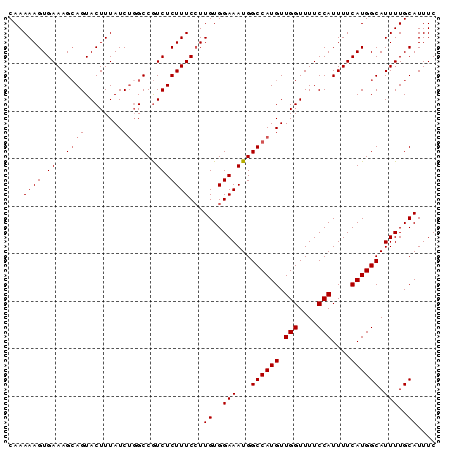
| Location | 13,283,178 – 13,283,297 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13283178 119 + 22224390 AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUUGAUUAUUUU-UU ..((((((....(((....))).))))))..(((((....)))))((((..(.((((((((.(((((((..(..................)..))))))).)))))))).)..))))-.. ( -29.77) >DroSec_CAF1 29362 118 + 1 AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCUGAUAAAGUACUGCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUUGAUUAUUU--UU ...((((....))))....(((((..((((.(((((....))))).....))))......((((..(((..........)))...)))).))))).(((((((.....))))))).--.. ( -27.60) >DroSim_CAF1 30067 118 + 1 AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCGAGAUAAAGUACUGCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUUGAUUAUUU--UU ..((((((....(((....))).))))))..(((((....)))))((((..(..(((((((.(((((((..(..................)..))))))).)))))))..)..)))--). ( -26.87) >DroEre_CAF1 29336 118 + 1 AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUCGAUUAUUU--UU ...((((....))))..........(((((.(((((....))))).....)))))((((((.(((((((..(..................)..))))))).)))))).........--.. ( -28.57) >DroYak_CAF1 29056 120 + 1 AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAUGAAAGAGAUGGCCAGAUAAAGUACUUCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUCGAUUAUUUUUUU .((((((((...(((((..((((..((((((((((..........))))))))))...(((((.........))))))))).))))).))).)))))((((((.....))))))...... ( -28.90) >consensus AUGCCAUGAAAAUGGAAAACCAACAUGGCCAUUUCCACAAGGAAAGAGACGGCCAGAUAAAGUACUGCUUUCACUUUUUGGCUUCUACUCGUUGGCAGUAAUUUAUUUGAUUAUUU__UU .((((((((...(((((..((((..(((((.(((((....))))).....)))))...((((.....))))......)))).))))).))).)))))((((((.....))))))...... (-24.20 = -24.80 + 0.60)

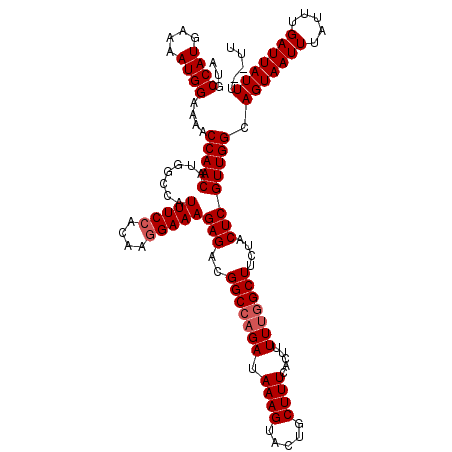

| Location | 13,283,178 – 13,283,297 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13283178 119 - 22224390 AA-AAAAUAAUCAAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU ..-....................(((((..((((.((((((((((((((.......))))))...(((((((....((((....)))))))))))..))))))))..))))...))))). ( -30.50) >DroSec_CAF1 29362 118 - 1 AA--AAAUAAUCAAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGCAGUACUUUAUCAGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU ..--...................(((((..((((.((((((((((((((.......))))))....((((((....((((....))))))))))...))))))))..))))...))))). ( -30.10) >DroSim_CAF1 30067 118 - 1 AA--AAAUAAUCAAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGCAGUACUUUAUCUCGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU ..--...................(((((..((((.((((((((...(((((((.....)))).....(((((....((((....)))))))))))).))))))))..))))...))))). ( -26.50) >DroEre_CAF1 29336 118 - 1 AA--AAAUAAUCGAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU ..--...................(((((..((((.((((((((((((((.......))))))...(((((((....((((....)))))))))))..))))))))..))))...))))). ( -30.50) >DroYak_CAF1 29056 120 - 1 AAAAAAAUAAUCGAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGAAGUACUUUAUCUGGCCAUCUCUUUCAUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU .......................(((((..((((.((((((((((((((.......))))))...(((((((.(((........))).)))))))..))))))))..))))...))))). ( -30.50) >consensus AA__AAAUAAUCAAAUAAAUUACUGCCAACGAGUAGAAGCCAAAAAGUGAAAGCAGUACUUUAUCUGGCCGUCUCUUUCCUUGUGGAAAUGGCCAUGUUGGUUUUCCAUUUUCAUGGCAU .......................(((((..((((.((((((((((((((.......))))))....((((((.(((........))).))))))...))))))))..))))...))))). (-28.48 = -28.52 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:42 2006