
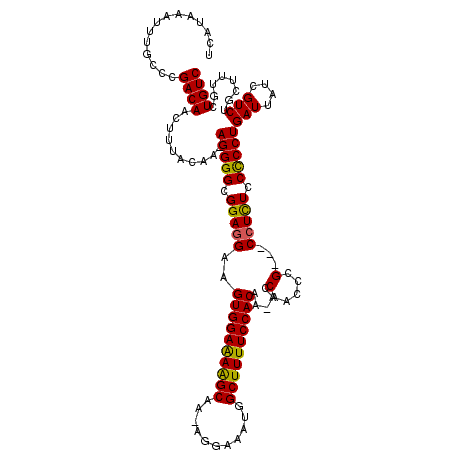
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,282,903 – 13,283,079 |
| Length | 176 |
| Max. P | 0.999909 |

| Location | 13,282,903 – 13,283,006 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 96.12 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.63 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13282903 103 + 22224390 AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGCCCGACAACUUUACAAAAGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAA ....((((.....))))...............(((((((................)))))))((..((((((((((..-........))))))))))..-.)).. ( -25.19) >DroSec_CAF1 29092 102 + 1 AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAA ....((((.....))))...............(((((((.............-..)))))))((..((((((((((..-........))))))))))..-.)).. ( -25.26) >DroSim_CAF1 29798 102 + 1 AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAA ....((((.....))))...............(((((((.............-..)))))))((..((((((((((..-........))))))))))..-.)).. ( -25.26) >DroEre_CAF1 29089 103 + 1 AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGGCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAAAACCAA ....((((.....))))................(((((((.(..(((....)-))..)))).....((((((((((..-........))))))))))....)))) ( -24.00) >DroYak_CAF1 28804 101 + 1 AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAUG--GUGGAGAGGCAAAAGGAAAUGGCUUUUCCACAA-GCCAA ....((((.....))))..................((((.............-..))))((...--((((((((((...........))))))))))..-.)).. ( -25.46) >consensus AAAAGAAAAACGAUUUCAAUAAAUGUCAUAAAUUUGCCCGACAACUUUACAA_AGGGGCGGAGGAAGUGGAAAAGCAA_AGGAAAUGGCUUUUCCACAA_ACCAA ....((((.....))))...............(((((((................)))))))....((((((((((...........))))))))))........ (-22.75 = -22.63 + -0.12)

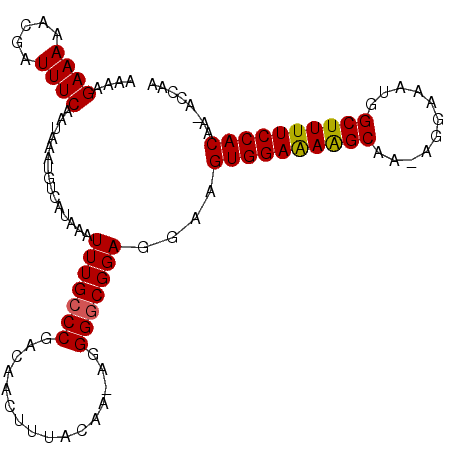

| Location | 13,282,903 – 13,283,006 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 96.12 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -19.51 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13282903 103 - 22224390 UUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCUUUUGUAAAGUUGUCGGGCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..((.-..((((((((((.(......-).))))))))))..))...((((................))))..................((((.....)))).... ( -23.19) >DroSec_CAF1 29092 102 - 1 UUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..((.-..((((((((((.(......-).))))))))))..))...((((..-.............))))..................((((.....)))).... ( -23.26) >DroSim_CAF1 29798 102 - 1 UUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..((.-..((((((((((.(......-).))))))))))..))...((((..-.............))))..................((((.....)))).... ( -23.26) >DroEre_CAF1 29089 103 - 1 UUGGUUUUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGCCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..((((((((((((((((.(......-).)))))))))).............-..(((((..(((((..........)))))))))).))))))........... ( -20.50) >DroYak_CAF1 28804 101 - 1 UUGGC-UUGUGGAAAAGCCAUUUCCUUUUGCCUCUCCAC--CAUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..(((-....(((((.....)))))....))).......--.....((((..-.............))))..................((((.....)))).... ( -16.06) >consensus UUGGU_UUGUGGAAAAGCCAUUUCCU_UUGCUUUUCCACUUCCUCCGCCCCU_UUGUAAAGUUGUCGGGCAAAUUUAUGACAUUUAUUGAAAUCGUUUUUCUUUU ..((....((((((((((...........)))))))))).....))((((................))))..................((((.....)))).... (-19.51 = -20.11 + 0.60)

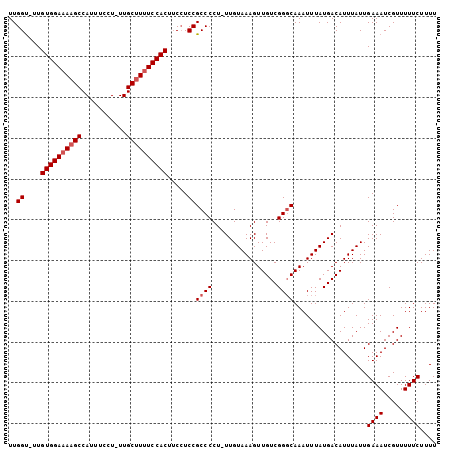
| Location | 13,282,928 – 13,283,046 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -28.54 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13282928 118 + 22224390 UCAUAAAUUUGCCCGACAACUUUACAAAAGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCGCCGCCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUC ..........((..(((...........(((((.(((((..((((((((((..-........))))))))))..-..(.....)...))))).)))))(.....)))).))......... ( -32.20) >DroSec_CAF1 29117 114 + 1 UCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCG---CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUC ..........((..(((..........-(((((.(((((..((((((((((..-........))))))))))..-.........---))))).)))))(.....)))).))......... ( -32.32) >DroSim_CAF1 29823 114 + 1 UCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCG---CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUC ..........((..(((..........-(((((.(((((..((((((((((..-........))))))))))..-.........---))))).)))))(.....)))).))......... ( -32.32) >DroEre_CAF1 29114 115 + 1 UCAUAAAUUUGGCCGACAACUUUACAA-AGGGGCGGAGGAAGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAAAACCAACCCG---CCUUUCCCCCUGAUUAUCGUCUGCUUUGCUGUC ..........(((.(((..........-(((((..((((..((((((((((..-........))))))))))............---))))..)))))(.....)))).)))........ ( -32.54) >DroYak_CAF1 28829 113 + 1 UCAUAAAUUUGCCCGACAACUUUACAA-AGGGGCGGAUG--GUGGAGAGGCAAAAGGAAAUGGCUUUUCCACAA-GCCAACCCG---CUUUUCCUCCUGAUUAUCGUCUGCUUUGCUGUC .......................(((.-.((((((((((--((((.((((.((((((...((((((......))-))))...).---)))))))))....))))))))))))))..))). ( -32.60) >consensus UCAUAAAUUUGCCCGACAACUUUACAA_AGGGGCGGAGGAAGUGGAAAAGCAA_AGGAAAUGGCUUUUCCACAA_ACCAACCCG___CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUC ..............((((..........(((((.(((((..((((((((((...........)))))))))).....(.....)...))))).)))))(((....)))........)))) (-28.54 = -28.02 + -0.52)


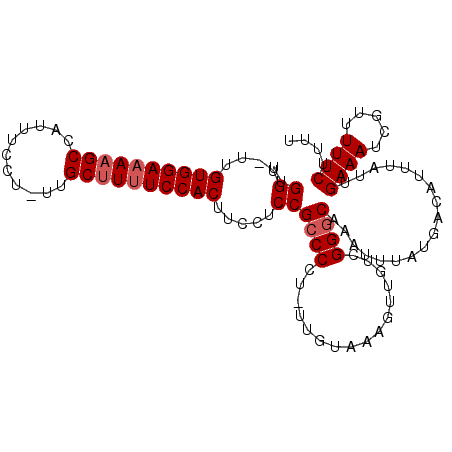
| Location | 13,282,928 – 13,283,046 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -40.00 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13282928 118 - 22224390 GACAGCAAAGCAGACGAUAAUCAGGGGGAGAGGCGGCGGGUUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCUUUUGUAAAGUUGUCGGGCAAAUUUAUGA .........((...(((((((....(.((((((.((((((..((.-..((((((((((.(......-).))))))))))..)))))))))))))).)...))))))).)).......... ( -45.60) >DroSec_CAF1 29117 114 - 1 GACAGCAAAGCAGACGAUAAUCAGGGGGAGAGG---CGGGUUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGA .........((...(((((((...(.((((.((---((((..((.-..((((((((((.(......-).))))))))))..)))))))).))-)).)...))))))).)).......... ( -41.80) >DroSim_CAF1 29823 114 - 1 GACAGCAAAGCAGACGAUAAUCAGGGGGAGAGG---CGGGUUGGU-UUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGA .........((...(((((((...(.((((.((---((((..((.-..((((((((((.(......-).))))))))))..)))))))).))-)).)...))))))).)).......... ( -41.80) >DroEre_CAF1 29114 115 - 1 GACAGCAAAGCAGACGAUAAUCAGGGGGAAAGG---CGGGUUGGUUUUGUGGAAAAGCCAUUUCCU-UUGCUUUUCCACUUCCUCCGCCCCU-UUGUAAAGUUGUCGGCCAAAUUUAUGA .........((.((((((...(((((((....(---((((..((....((((((((((.(......-).))))))))))..)))))))))))-)))....)))))).))........... ( -40.10) >DroYak_CAF1 28829 113 - 1 GACAGCAAAGCAGACGAUAAUCAGGAGGAAAAG---CGGGUUGGC-UUGUGGAAAAGCCAUUUCCUUUUGCCUCUCCAC--CAUCCGCCCCU-UUGUAAAGUUGUCGGGCAAAUUUAUGA ((((((((((..(..(((.....((((((((((---.(((.((((-((......)))))).)))))))).)))))....--.)))..)..))-)))).....)))).............. ( -30.70) >consensus GACAGCAAAGCAGACGAUAAUCAGGGGGAGAGG___CGGGUUGGU_UUGUGGAAAAGCCAUUUCCU_UUGCUUUUCCACUUCCUCCGCCCCU_UUGUAAAGUUGUCGGGCAAAUUUAUGA ((((((...((((.......((.....))........(((.(((....((((((((((...........)))))))))).....))).)))..))))...)))))).............. (-29.50 = -29.90 + 0.40)

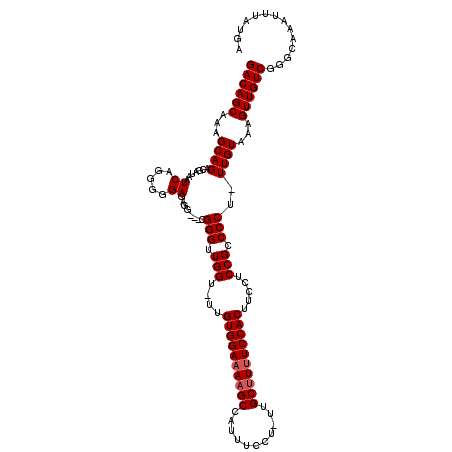
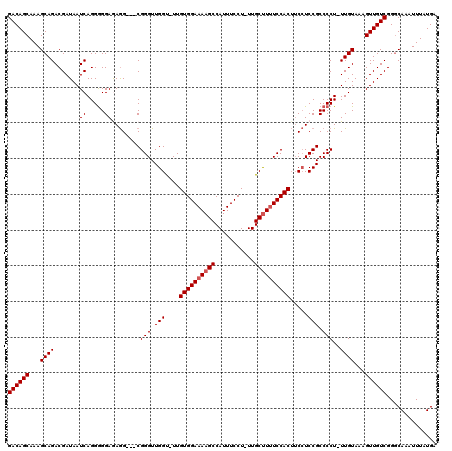
| Location | 13,282,968 – 13,283,079 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13282968 111 + 22224390 AGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCGCCGCCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUCGGCUUGUAAUCGCUUUCAUUACAGCAG-------AAAAAG .((((((((((..-........))))))))))..-...............................((((((.((.((..(((........)))..)).)).)))))-------)..... ( -27.50) >DroSec_CAF1 29156 108 + 1 AGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCG---CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUCGGCUUGUAAUCGCUUUCAUUACAGCAG-------AAAAAG .((((((((((..-........))))))))))..-.........---...................((((((.((.((..(((........)))..)).)).)))))-------)..... ( -27.50) >DroSim_CAF1 29862 108 + 1 AGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAA-ACCAACCCG---CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUCGGCUUGUAAUCGCUUUCAUUACAGCAG-------AAAAAG .((((((((((..-........))))))))))..-.........---...................((((((.((.((..(((........)))..)).)).)))))-------)..... ( -27.50) >DroEre_CAF1 29153 109 + 1 AGUGGAAAAGCAA-AGGAAAUGGCUUUUCCACAAAACCAACCCG---CCUUUCCCCCUGAUUAUCGUCUGCUUUGCUGUCGGCGUGUAAUCGCUUUCAUUACAGCAG-------AAAAAG .((((((((((..-........))))))))))............---...................((((((.((.((..((((......))))..)).)).)))))-------)..... ( -30.20) >DroYak_CAF1 28867 115 + 1 -GUGGAGAGGCAAAAGGAAAUGGCUUUUCCACAA-GCCAACCCG---CUUUUCCUCCUGAUUAUCGUCUGCUUUGCUGUCGGCUUGUAAUCGCUUUCAUUACAGCAGCAAAAAAAAAAAG -..((((((((....((...((((((......))-))))..)))---)))))))...........(.(((((.((.((..(((........)))..)).)).))))))............ ( -29.30) >consensus AGUGGAAAAGCAA_AGGAAAUGGCUUUUCCACAA_ACCAACCCG___CCUCUCCCCCUGAUUAUCGUCUGCUUUGCUGUCGGCUUGUAAUCGCUUUCAUUACAGCAG_______AAAAAG .((((((((((...........))))))))))...................................(((((.((.((..(((........)))..)).)).)))))............. (-25.04 = -24.72 + -0.32)

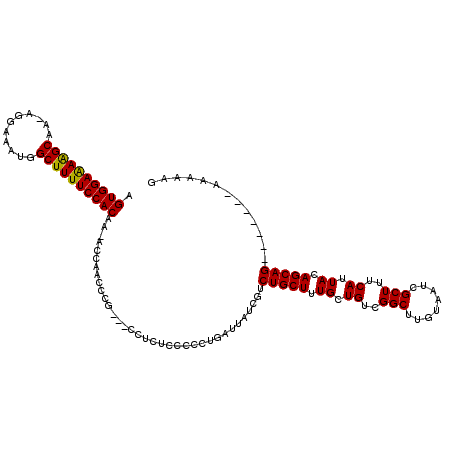
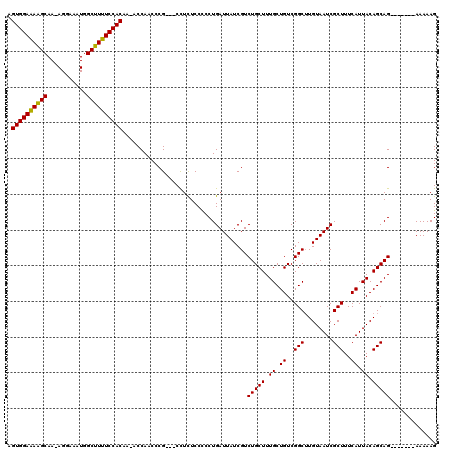
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:38 2006