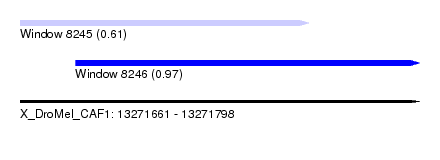
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,271,661 – 13,271,798 |
| Length | 137 |
| Max. P | 0.974093 |

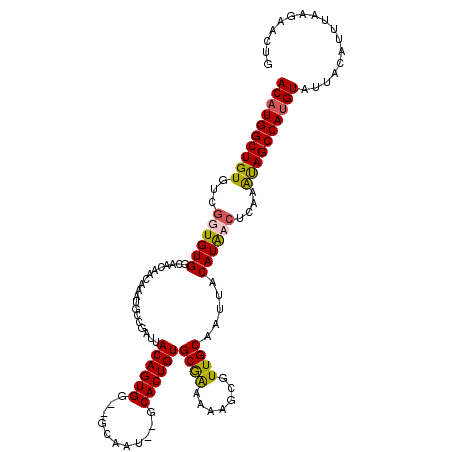
| Location | 13,271,661 – 13,271,760 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -21.33 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13271661 99 + 22224390 CUCACACAAAGUGGUUAA-AACAUGGCUGUGUCAGUGUGGCAACAACAAAUGGCGAUUACAGUGGUCACAAUGGGCACUGUGCGAAAAAGGAUUGCAAUU .((((((...(..((((.-....))))..)....))))))............((((((((((((.((.....)).)))))).(......))))))).... ( -24.00) >DroSec_CAF1 16132 95 + 1 CUCACACAAAGUGGUUAA-AACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGC--GCAAU--GCACUGUGCGAAAAAGCGUUGCAAUU ..(((((...(..((((.-....))))..)....)))))(((((..............(((((((--.....--)))))))((......))))))).... ( -30.20) >DroSim_CAF1 16605 95 + 1 CUCACACAAAGUGGUUAA-AACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGG--GCAAU--GCACUGUGCGAAAAAGCGUUGCAAUU ..(((((...(..((((.-....))))..)....)))))(((((......((((.((....)).)--)))..--.......((......))))))).... ( -27.00) >DroEre_CAF1 17998 96 + 1 CUCACACAAAGUGGUUAAAAACAUGGCUGUGUCGGUGUGGCAACAACAAAUGACGACUACAGUGG--GCAAU--GCACUGUGCAGAAAAACGUUGCCAUU ((.(((((..(((........)))...))))).)).((((((((..(....).....(((((((.--.....--.))))))).........)))))))). ( -25.00) >consensus CUCACACAAAGUGGUUAA_AACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGG__GCAAU__GCACUGUGCGAAAAAGCGUUGCAAUU ..(((((...(..((((......))))..)....)))))(((((..............((((((...........))))))((......))))))).... (-21.33 = -22.07 + 0.75)

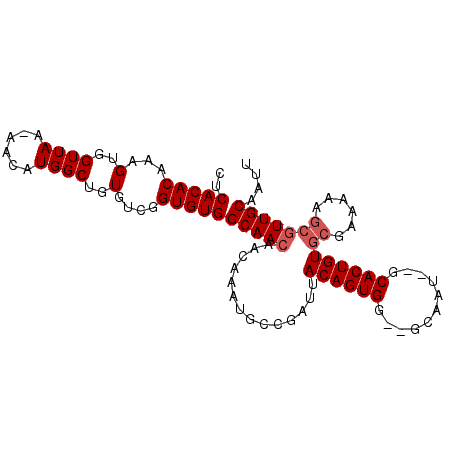

| Location | 13,271,680 – 13,271,798 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -27.49 |
| Energy contribution | -27.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13271680 118 + 22224390 ACAUGGCUGUGUCAGUGUGGCAACAACAAAUGGCGAUUACAGUGGUCACAAUGGGCACUGUGCGAAAAAGGAUUGCAAUUACAUACCUCAAAUAGCCAUGUAUUACAUUUAAGAACUG ((((((((((((((...((....)).....))))((..((((((.((.....)).))))))((((.......))))...........))..))))))))))................. ( -33.50) >DroSec_CAF1 16151 114 + 1 ACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGC--GCAAU--GCACUGUGCGAAAAAGCGUUGCAAUUACAUAACUCAAAUAGCCAUGUAUUACAUUUAAGAACUG ((((((((((....((((((((((..............(((((((--.....--)))))))((......)))))))...))))).......))))))))))................. ( -36.50) >DroSim_CAF1 16624 114 + 1 ACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGG--GCAAU--GCACUGUGCGAAAAAGCGUUGCAAUUGCAUAACUCAAAUAGCCAUGUAUUACAUUUAAGAACUG ((((((((((((((((.((....))......)))))).....(((--(..((--(((...(((((.......)))))..)))))..)))).))))))))))................. ( -36.00) >DroEre_CAF1 18018 114 + 1 ACAUGGCUGUGUCGGUGUGGCAACAACAAAUGACGACUACAGUGG--GCAAU--GCACUGUGCAGAAAAACGUUGCCAUUACAUGCCUCGAGCAGCCAAGUAAAGCUGGUAAGAACUC ((.(((((((...(((((((((((..(....).....(((((((.--.....--.))))))).........)))))))).....)))....))))))).))................. ( -33.20) >consensus ACAUGGCUGUGUCGGUGUGGCAACAACAAAUGCCGAUUACAGUGG__GCAAU__GCACUGUGCGAAAAAGCGUUGCAAUUACAUAACUCAAAUAGCCAUGUAUUACAUUUAAGAACUG ((((((((((...((((((...................((((((...........))))))((((.......)))).....))))))....))))))))))................. (-27.49 = -27.55 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:25 2006