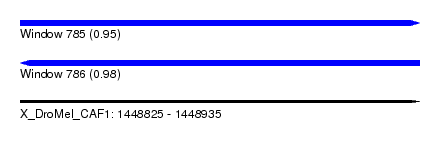
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,448,825 – 1,448,935 |
| Length | 110 |
| Max. P | 0.982977 |

| Location | 1,448,825 – 1,448,935 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.24 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1448825 110 + 22224390 GCCGCGUUUACUUAGUUAGUCCGAAA-GAGGCAGAGAAACAAGAGGAGCGAGGGGGGGGGGGGAA------AAGUCGCGGAUUGCGACAGCUCCAAAAUGCGGCGACAAAGGUAAAA (((((((((.........((((....-).)))............(((((................------..(((((.....))))).))))).)))))))))............. ( -33.61) >DroSim_CAF1 35799 106 + 1 GCCGCGUUUACUUAGUUAGUCCGAAA-AAGGGAGAGAAACGAGAGGAGCGAGGGA----GAGGAA------AAGUCGCGGAUUGCGACAGCUCCAAAAUGCGGCGACAAAGGUAAAA (((((((((.(((..((..(((....-..)))....))..))).(((((......----......------..(((((.....))))).))))).)))))))))............. ( -32.66) >DroAna_CAF1 65572 113 + 1 GCCGCGUUUACUUAGUUAGUCCGGACGGACAAAAAGAGAUAGGUGGAGAGAGGGG----AGGGAAUACCCGGAGUCGCGGAUUGCGACAGCUCCGAAAUGCCGCGACAAAGGUAAAA ((.((((((.(((..((.((((....)))).))..))).....(((((...(((.----........)))...(((((.....)))))..))))))))))).))............. ( -35.00) >consensus GCCGCGUUUACUUAGUUAGUCCGAAA_GAGGAAGAGAAACAAGAGGAGCGAGGGG____GGGGAA______AAGUCGCGGAUUGCGACAGCUCCAAAAUGCGGCGACAAAGGUAAAA ((((((((((((.....)))........................(((((........................(((((.....))))).))))).)))))))))............. (-23.58 = -24.24 + 0.67)

| Location | 1,448,825 – 1,448,935 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -19.38 |
| Energy contribution | -20.04 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1448825 110 - 22224390 UUUUACCUUUGUCGCCGCAUUUUGGAGCUGUCGCAAUCCGCGACUU------UUCCCCCCCCCCCCUCGCUCCUCUUGUUUCUCUGCCUC-UUUCGGACUAACUAAGUAAACGCGGC .............(((((.....(((((.(((((.....)))))..------................))))).((((((..((((....-...))))..))).))).....))))) ( -27.41) >DroSim_CAF1 35799 106 - 1 UUUUACCUUUGUCGCCGCAUUUUGGAGCUGUCGCAAUCCGCGACUU------UUCCUC----UCCCUCGCUCCUCUCGUUUCUCUCCCUU-UUUCGGACUAACUAAGUAAACGCGGC .............(((((.....(((((.(((((.....)))))..------......----......)))))....((((.....((..-....))(((.....)))))))))))) ( -25.36) >DroAna_CAF1 65572 113 - 1 UUUUACCUUUGUCGCGGCAUUUCGGAGCUGUCGCAAUCCGCGACUCCGGGUAUUCCCU----CCCCUCUCUCCACCUAUCUCUUUUUGUCCGUCCGGACUAACUAAGUAAACGCGGC ..........((((((.......((((..(((((.....)))))...(((....))).----.......))))..............((((....))))............)))))) ( -32.10) >consensus UUUUACCUUUGUCGCCGCAUUUUGGAGCUGUCGCAAUCCGCGACUU______UUCCCC____CCCCUCGCUCCUCUUGUUUCUCUCCCUC_UUUCGGACUAACUAAGUAAACGCGGC .............(((((.....(((((.(((((.....)))))........................)))))......................(.(((.....)))...)))))) (-19.38 = -20.04 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:17 2006