| Sequence ID | X_DroMel_CAF1 |
|---|---|
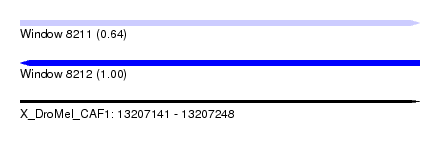
| Location | 13,207,141 – 13,207,248 |
| Length | 107 |
| Max. P | 0.998297 |

| Location | 13,207,141 – 13,207,248 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -16.68 |
| Energy contribution | -18.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13207141 107 + 22224390 ----------AGUUGCUUGUGCAAAGCCCCAAAACUCUGAUAAGAAAUGCUCGAUUGCAGUUUUGAAAUGCGCGCCUUGCGGCGAGUGCAAUUGGGUUUGAGUCUAGGAUAAAUGCA-- ----------.(((.((((.((.(((((((((((((((....)))..(((......)))))))))...((((((((....)))..)))))...))))))..)).))))...)))...-- ( -29.40) >DroSec_CAF1 35443 84 + 1 -----------------------------------UUUGAUAAGAAAUGCUCGAAUGCAGUUUUGAAAUGCGCGCCUUGCGGCGAGUGCAUUUGGGUUUGAGUCUAGGCUAAAUGCACC -----------------------------------.......(((...(((((((((((((........)).((((....))))..))))))))))).....)))..((.....))... ( -23.70) >DroSim_CAF1 36217 108 + 1 ----------AGUUGUUUGUGCAAAGCCC-GAAACUCUGAUAAGAAAUGCUCGAAUGCAGUUUUGAAAUGCGCGCCUUGCGGCGAUUGCAUUUGGGUUUGAGCCUAGGCUAAAUGCACC ----------........(((((((((((-((((((((....)))..(((......)))))))).((((((.((((....))))...)))))))))))).(((....)))...))))). ( -36.50) >DroEre_CAF1 44992 117 + 1 UCGUAAACUUUGUUGUUUUUGGAAAACCG-AAAACUCAGAUACGAAAUGCUUGAUUGCACUUUUAAAGUGCGCGCCUUGCGGCGAGUGCAUUU-GGUUUGAGUCUAGGCUAAAUGCACA (((((...((((..((((((((....)))-))))).)))))))))...........(((((.....))))).((((....)))).((((((((-((((((....)))))))))))))). ( -43.20) >DroYak_CAF1 44088 103 + 1 ---------------AUUUUGCAGUAUCG-GAAACUCAGAUAAGAAAUGCAUAGUUGCAGUUUUGAAAUGCGCGCCUUGCGACGAGUGCAUCGGGGUUUGAGUCCAGGCUAAAUGCACA ---------------....((((.(((((-....)...)))).....))))..(((((((................)))))))..((((((...((((((....))))))..)))))). ( -26.19) >consensus ___________GUUGUUUGUGCAAAACCC_AAAACUCUGAUAAGAAAUGCUCGAUUGCAGUUUUGAAAUGCGCGCCUUGCGGCGAGUGCAUUUGGGUUUGAGUCUAGGCUAAAUGCACA ..........................................((((.(((......))).))))........((((....)))).((((((((.((((((....)))))))))))))). (-16.68 = -18.12 + 1.44)



| Location | 13,207,141 – 13,207,248 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -17.65 |
| Energy contribution | -19.49 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13207141 107 - 22224390 --UGCAUUUAUCCUAGACUCAAACCCAAUUGCACUCGCCGCAAGGCGCGCAUUUCAAAACUGCAAUCGAGCAUUUCUUAUCAGAGUUUUGGGGCUUUGCACAAGCAACU---------- --((((....(((.((((((.........(((...((((....)))).))).........(((......)))..........)))))).)))....)))).........---------- ( -28.90) >DroSec_CAF1 35443 84 - 1 GGUGCAUUUAGCCUAGACUCAAACCCAAAUGCACUCGCCGCAAGGCGCGCAUUUCAAAACUGCAUUCGAGCAUUUCUUAUCAAA----------------------------------- (((((((((.................)))))))))((((....)))).............(((......)))............----------------------------------- ( -18.63) >DroSim_CAF1 36217 108 - 1 GGUGCAUUUAGCCUAGGCUCAAACCCAAAUGCAAUCGCCGCAAGGCGCGCAUUUCAAAACUGCAUUCGAGCAUUUCUUAUCAGAGUUUC-GGGCUUUGCACAAACAACU---------- .(((((...(((((((((((......((((((...((((....)))).))))))......(((......)))..........)))))).-))))).)))))........---------- ( -38.40) >DroEre_CAF1 44992 117 - 1 UGUGCAUUUAGCCUAGACUCAAACC-AAAUGCACUCGCCGCAAGGCGCGCACUUUAAAAGUGCAAUCAAGCAUUUCGUAUCUGAGUUUU-CGGUUUUCCAAAAACAACAAAGUUUACGA (((......((((.(((((((.((.-((((((...((((....)))).(((((.....)))))......)))))).))...))))))).-.))))........)))............. ( -34.14) >DroYak_CAF1 44088 103 - 1 UGUGCAUUUAGCCUGGACUCAAACCCCGAUGCACUCGUCGCAAGGCGCGCAUUUCAAAACUGCAACUAUGCAUUUCUUAUCUGAGUUUC-CGAUACUGCAAAAU--------------- ..((((.....(..(((((((......(((((...((((....)))).))))).......((((....)))).........))))))).-.)....))))....--------------- ( -23.70) >consensus GGUGCAUUUAGCCUAGACUCAAACCCAAAUGCACUCGCCGCAAGGCGCGCAUUUCAAAACUGCAAUCGAGCAUUUCUUAUCAGAGUUUC_CGGCUUUGCAAAAACAAC___________ ..((((........((((((......((((((...((((....)))).(((.........)))......)))))).......))))))........))))................... (-17.65 = -19.49 + 1.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:53 2006