
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,199,483 – 13,199,583 |
| Length | 100 |
| Max. P | 0.799202 |

| Location | 13,199,483 – 13,199,583 |
|---|---|
| Length | 100 |
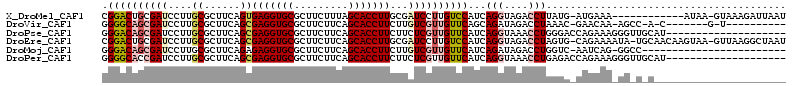
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.81 |
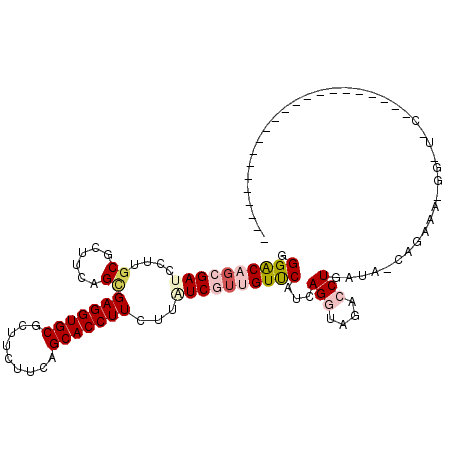
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13199483 100 - 22224390 CGGACUGCGAUCCUUGCGCUUCAGUGAGGUGCGCUUCUUUAGCACCUUGCGAUCCUUGUCCAUCAGGUAGACCUUAUG-AUGAAA------------AUAA-GUAAAGAUUAAU .(((.(((((...))))).))).(..((((((.........))))))..)(((((((((.(((((((....))...))-)))...------------))))-)....))))... ( -26.40) >DroVir_CAF1 24400 92 - 1 GGGGCAGCGAUCCUUGCGCUUCAGCGAGGUGCGCUUCUUCAGCACCUUCUUGUCGUUGUUCAGCAGAUAGACCUAAAC-GAACAA-AGCC-A-C-------G-U---------- ..(((((((..((((((......))))))..))))..............((((((((..((........))....)))-).))))-.)))-.-.-------.-.---------- ( -25.60) >DroPse_CAF1 22230 94 - 1 GGGACAGCGAUCCUUGCGCUUCAGCGAGGUGCGCUUCUUCAGCACCUUCUUCUCGUUGUUCAUCAGGUAAACCUGGGACCAGAAAGGGUUGCAU-------------------- ((((.((((..((((((......))))))..))))))))..(((((((.((((.(((.....(((((....)))))))).)))))))).)))..-------------------- ( -34.00) >DroEre_CAF1 37320 111 - 1 CGGACUGCGAUCCUUGCGCUUCAGCGAGGUGCGCUUCUUCAGCACCUUGCGAUCCUUGUCCAUCAGGUAGACCUAGUG-CAGAAAAUA-UGCAACAAGUAA-GUUAAGGCUAAU .((((.(((.......)))....(((((((((.........))))))))).......))))......(((.(((...(-((.......-)))(((......-))).)))))).. ( -32.70) >DroMoj_CAF1 18694 88 - 1 GGGACAGCGAUCCUUGCGCUUCAGAGAGGUGCGCUUCUUCAGCACCUUCUUGUCGUUGUUCAUCAGAUAGACCUGGUC-AAUCAG-GGCC------------------------ .((((((((((....(((((((...)))))))(((.....)))........))))))))))..........(((((..-..))))-)...------------------------ ( -29.20) >DroPer_CAF1 23383 94 - 1 GGGGCACCGAUCCUUGCGCUUCAGCGAGGUGCGCUUCUUCAGCACCUUCUUCUCGUUGUUCAUCAGGUAAACCUGAGACCAGAAAGGGUUGCAU-------------------- (((((.(....((((((......)))))).).)))))....(((((((.((((.(((.....(((((....)))))))).)))))))).)))..-------------------- ( -32.70) >consensus GGGACAGCGAUCCUUGCGCUUCAGCGAGGUGCGCUUCUUCAGCACCUUCUUAUCGUUGUUCAUCAGGUAGACCUGAUA_CAGAAA_GG_U_C______________________ .((((((((((....((......))(((((((.........)))))))...))))))))))...(((....)))........................................ (-20.80 = -21.80 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:49 2006