
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,191,827 – 13,191,956 |
| Length | 129 |
| Max. P | 0.938749 |

| Location | 13,191,827 – 13,191,921 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -24.15 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13191827 94 + 22224390 AUAUAUGCCUUUCGCCUUUUUGGCAUUUC-GGCAGUGC-CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUU .........((((((.(((((((((((..-...)))))-))))))(((((.....((((((.........))))))....)))))...)))))).. ( -30.80) >DroSim_CAF1 22797 88 + 1 GUGG-----AUUCG-CUUUUUGGCAUUUC-GGCAGUGC-CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUU ....-----.((((-((((((((((((..-...)))))-))))))(((((.....((((((.........))))))....)))))...)))))... ( -30.70) >DroEre_CAF1 30714 90 + 1 AUAU-----GUUCG-CUUUUUGGCAUUUUGGGCAGUGCCCAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUU ....-----.((((-(.((((.(((((((((((...))))))))..)))(((((.((((((.........))))))..))))))))).)))))... ( -31.50) >DroYak_CAF1 29369 89 + 1 AUAU-----GUACG-CUUUUUGGCAUUUUGGGCAGUGC-CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUU ....-----...((-((((((((((((......)))))-))))))(((((.....((((((.........))))))....)))))...)))..... ( -28.60) >consensus AUAU_____GUUCG_CUUUUUGGCAUUUC_GGCAGUGC_CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUU ..................(((.(((((((..(((((........)))))(((((.((((((.........))))))..)))))))))))).))).. (-24.15 = -24.65 + 0.50)



| Location | 13,191,827 – 13,191,921 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13191827 94 - 22224390 AAUUUCACAUUUUGCAACUUGCCGCAUUUUGAUUUUGCGGCCAUUACGCAAUUUUUG-GCACUGCC-GAAAUGCCAAAAAGGCGAAAGGCAUAUAU ............(((.....((((((.........)))))).....(((..((((((-(((.(...-..).))))))))).)))....)))..... ( -26.50) >DroSim_CAF1 22797 88 - 1 AAUUUCACAUUUUGCAACUUGCCGCAUUUUGAUUUUGCGGCCAUUACGCAAUUUUUG-GCACUGCC-GAAAUGCCAAAAAG-CGAAU-----CCAC ..........(((((.....((((((.........))))))..........((((((-(((.(...-..).))))))))))-)))).-----.... ( -23.90) >DroEre_CAF1 30714 90 - 1 AAUUUCACAUUUUGCAACUUGCCGCAUUUUGAUUUUGCGGCCAUUACGCAAUUUUUGGGCACUGCCCAAAAUGCCAAAAAG-CGAAC-----AUAU ..........(((((.....((((((.........))))))......(((..((((((((...)))))))))))......)-)))).-----.... ( -26.50) >DroYak_CAF1 29369 89 - 1 AAUUUCACAUUUUGCAACUUGCCGCAUUUUGAUUUUGCGGCCAUUACGCAAUUUUUG-GCACUGCCCAAAAUGCCAAAAAG-CGUAC-----AUAU ....................((((((.........))))))...(((((..((((((-(((.(......).))))))))))-)))).-----.... ( -25.10) >consensus AAUUUCACAUUUUGCAACUUGCCGCAUUUUGAUUUUGCGGCCAUUACGCAAUUUUUG_GCACUGCC_AAAAUGCCAAAAAG_CGAAC_____AUAU ...........((((.....((((((.........))))))......))))((((((.(((.(......).)))))))))................ (-18.50 = -18.50 + 0.00)

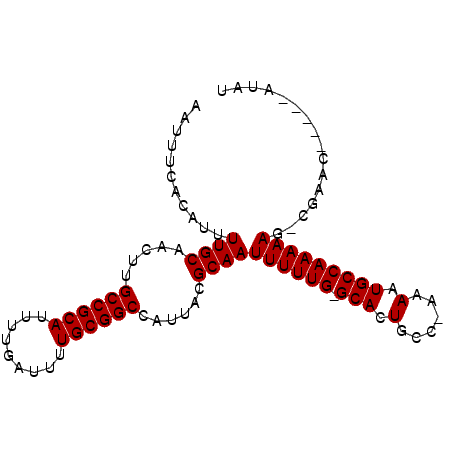

| Location | 13,191,864 – 13,191,956 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.84 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13191864 92 + 22224390 -CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUUUAUUAGGCGCUUCGGGGCAACGAGAGAGCGAGAGA -......(((((((((.((((((.........))))))..)))))...................((.(....).)).........)))).... ( -23.80) >DroSim_CAF1 22828 92 + 1 -CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUUUAUUAGGCGCUUCGGGGCUUCGAGAGAGCGAGAGA -......(((((((((.((((((.........))))))..)))))..................(((.(....).)))((....)))))).... ( -25.20) >DroEre_CAF1 30746 93 + 1 CCAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUUUAUUAGGCGCUUCGGGGCUUCGAGAGAGCGAGAGA .......(((((((((.((((((.........))))))..)))))..................(((.(....).)))((....)))))).... ( -25.20) >consensus _CAAAAAUUGCGUAAUGGCCGCAAAAUCAAAAUGCGGCAAGUUGCAAAAUGUGAAAUUUAUUAGGCGCUUCGGGGCUUCGAGAGAGCGAGAGA .........((.(((((((((((.........))))))...(..(.....)..)....))))).))((((((......)))...)))...... (-21.40 = -21.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:48 2006