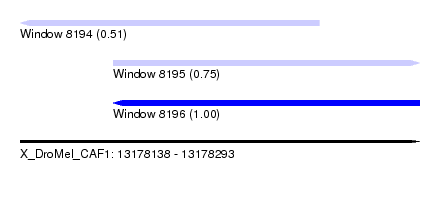
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,178,138 – 13,178,293 |
| Length | 155 |
| Max. P | 0.996463 |

| Location | 13,178,138 – 13,178,254 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13178138 116 - 22224390 CCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUUUACCUGAGAGCAUUUGG----CGUAGGCUAGAUUU .....(((((.((((((.....))))))...)))))((((....((((((((....))))))))((.((.....)).)).............))))((((((----(....))))))).. ( -27.40) >DroSec_CAF1 8087 116 - 1 CCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUUUACCUGAGAGCAUUUGA----CGUAGACUAGAUUU .....(((((.((((((.....))))))...)))))(((((((.((((((((....))))).((((.((.....)).))))............))).)))))----.))........... ( -22.30) >DroSim_CAF1 8209 116 - 1 CCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUUUACCUGAGAGCAUUUGG----CGUAGGCUAGAUUU .....(((((.((((((.....))))))...)))))((((....((((((((....))))))))((.((.....)).)).............))))((((((----(....))))))).. ( -27.40) >DroEre_CAF1 12063 120 - 1 CCCAACAUGUUUGUAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUUUACCUGAGAGCAUUUGGCCCUCGUAGGCUAGAUUU .....(((((.((((((.....))))))...)))))((((....((((((((....))))))))((.((.....)).)).............))))((((((((......)))))))).. ( -28.30) >DroYak_CAF1 13819 116 - 1 CCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUCUACCUGAGAGCAUUCGG----CGUAGGCUAGAUUU .....(((((.((((((.....))))))...)))))(((((...(((((.............((((.((.....)).)))).((((.....))))...))))----).)))))....... ( -28.70) >consensus CCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACACAUUUUACCUGAGAGCAUUUGG____CGUAGGCUAGAUUU .....(((((.((((((.....))))))...)))))(((((...((((((((....))))))))(((.((...((............)).)).)))............)))))....... (-23.16 = -23.20 + 0.04)


| Location | 13,178,174 – 13,178,293 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -18.36 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13178174 119 + 22224390 UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUGCAAACAUGUUGGGUAAA-AGGAAACAAGGAAACAAAAAGAAAAAAAAAGAUGU (((..((..........((.((((....)))).))((((((..((((((((..(((((.....)))))..))))).))).))))-)).......))..)))................... ( -19.20) >DroSec_CAF1 8123 118 + 1 UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUGCAAACAUGUUGGGUAAA-AGGAAACAAGGAAACAAAACAAAAA-AAAAUAUGU (((..((..........((.((((....)))).))((((((..((((((((..(((((.....)))))..))))).))).))))-)).......))..))).........-......... ( -19.20) >DroSim_CAF1 8245 119 + 1 UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUGCAAACAUGUUGGGUAAA-AGGAAACAAGGAAACAAAACAAAAAAAAAAGAUGU (((..((..........((.((((....)))).))((((((..((((((((..(((((.....)))))..))))).))).))))-)).......))..)))................... ( -19.20) >DroEre_CAF1 12103 109 + 1 UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUACAAACAUGUUGGGAAAG--GGAAACAAGGAAACAA---------AAAAGAUGU (((..((.....(((((((.....))........(..(((((...((((....))))...)))))..).....)))))......--))..))).(....)..---------......... ( -14.90) >DroYak_CAF1 13855 111 + 1 UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUGCAAACAUGUUGGGAAAAGAGGAAACCAGGAAACAA---------AAAAGAUGU (((..((..((......))......((....((..(((((...((((((((..(((((.....)))))..))))).))).)))))..))..)).))..))).---------......... ( -19.30) >consensus UGUGGCCAAAGACAACACUAAUUAAGGAUAAUCAGCUUUUAAGCCAUGUGUACGCAUUAAUUAAAUGCAAACAUGUUGGGUAAA_AGGAAACAAGGAAACAAAA__AAAA_AAAAGAUGU (((..((.....((((..................(((....)))...((((..(((((.....)))))..))))))))........(....)..))..)))................... (-17.08 = -17.28 + 0.20)


| Location | 13,178,174 – 13,178,293 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.04 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13178174 119 - 22224390 ACAUCUUUUUUUUUCUUUUUGUUUCCUUGUUUCCU-UUUACCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA ...................................-...........(((.((((((.....)))))).)))..(((((.....((((((((....)))))))).........))))).. ( -19.44) >DroSec_CAF1 8123 118 - 1 ACAUAUUUU-UUUUUGUUUUGUUUCCUUGUUUCCU-UUUACCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA .........-.........................-...........(((.((((((.....)))))).)))..(((((.....((((((((....)))))))).........))))).. ( -19.44) >DroSim_CAF1 8245 119 - 1 ACAUCUUUUUUUUUUGUUUUGUUUCCUUGUUUCCU-UUUACCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA ...................................-...........(((.((((((.....)))))).)))..(((((.....((((((((....)))))))).........))))).. ( -19.44) >DroEre_CAF1 12103 109 - 1 ACAUCUUUU---------UUGUUUCCUUGUUUCC--CUUUCCCAACAUGUUUGUAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA .........---------................--...........(((.((((((.....)))))).)))..(((((.....((((((((....)))))))).........))))).. ( -17.44) >DroYak_CAF1 13855 111 - 1 ACAUCUUUU---------UUGUUUCCUGGUUUCCUCUUUUCCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA .........---------........(((((..............(((((.((((((.....))))))...)))))........((((((((....)))))))).........))))).. ( -19.90) >consensus ACAUCUUUU_UUUU__UUUUGUUUCCUUGUUUCCU_UUUACCCAACAUGUUUGCAUUUAAUUAAUGCGUACACAUGGCUUAAAAGCUGAUUAUCCUUAAUUAGUGUUGUCUUUGGCCACA ...............................................(((.((((((.....)))))).)))..(((((.....((((((((....)))))))).........))))).. (-19.20 = -19.04 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:38 2006