| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,177,219 – 13,177,329 |
| Length | 110 |
| Max. P | 0.832664 |

| Location | 13,177,219 – 13,177,329 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -47.07 |
| Consensus MFE | -33.53 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13177219 110 - 22224390 UCUCUGGCCGCACGGCCAUGGCUGCAAUGGCGGCAAUGCAUUGCAAUGGC--CUGGCCUGGCUGGUCUGCAGGAGGCCCAGAUUGAAUCGCUGGCUUUUUGGCAAGGGCCAA ((((.((((((..(((((.((((((((((.((....))))))))...)))--))))))..)).)))).).))).(((((.(((...)))((..(....)..))..))))).. ( -46.70) >DroSec_CAF1 7221 93 - 1 UCUCUGGCCGGACGGCCGUGGC---------UGCAA----------UGGCUGCUGGCCUGGCUGGUCUGCAGGAGGCCCAGAUUGAAUCGCUUGCUUUUUGGCAAGGGCCAA .....((((((.(((((((...---------....)----------))))))))))))((((((((((.....)))))............((((((....))))))))))). ( -42.70) >DroSim_CAF1 7336 102 - 1 UCUCUGGCCGGACGGCCAUGGCUGCACUGGCUGCAA----------UGGCUGCUGGCCUGGCUGGUCUGCAGGAGGCCCAGAUUGAAUCGCUGGCUUUUUGGCAAGGGCCAA .....((((((.(((((((.((.((....)).)).)----------))))))))))))....((((((.(((((((((..(((...)))...)))))))))....)))))). ( -47.80) >DroEre_CAF1 11293 107 - 1 UCACUGGCCGGAUGGCCAUGGCUGCCAUGGCUGCCCUGG-CUGCCCUGG---CUGCCCUGGCUGG-CUGCAGCAGGCCCAGAUUGAAUCGCUGGCUUUUUGGCAAGGGCCAA .....((((....(((((.(((.((((.(((.((....)-).))).)))---).))).)))))))-))......(((((.(((...)))((..(....)..))..))))).. ( -51.10) >consensus UCUCUGGCCGGACGGCCAUGGCUGCAAUGGCUGCAA__________UGGC__CUGGCCUGGCUGGUCUGCAGGAGGCCCAGAUUGAAUCGCUGGCUUUUUGGCAAGGGCCAA ....(((((...((((((..((.((....)).)).............(((.....)))))))))...(((((((((((..(((...)))...)))))))).)))..))))). (-33.53 = -34.02 + 0.50)

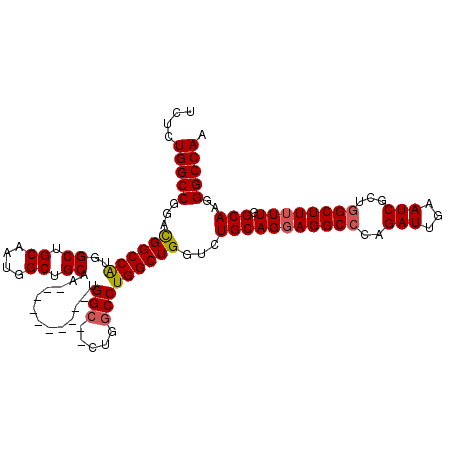

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:35 2006