| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,437,046 – 1,437,147 |
| Length | 101 |
| Max. P | 0.973443 |

| Location | 1,437,046 – 1,437,147 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -44.06 |
| Consensus MFE | -23.02 |
| Energy contribution | -26.19 |
| Covariance contribution | 3.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1437046 101 + 22224390 CGCGGAGUGGGCGGA-----------AAAUGGGCGGGGGACUAGUGCUUGGCUUUGUGC-UGGGCCUACAAUACAGUGACUGCCACCCACCUUCC-------CCCGCCCACAGCCCCUUG ....(((.((((...-----------...(((((((((((...(((..((((...((((-((...........)))).)).))))..)))..)))-------))))))))..))))))). ( -48.80) >DroSec_CAF1 41766 112 + 1 CGCGGAGUGGGCGGAAAGUGGGCGGAAAGUGGGCGGGGGACUAGUGCUUGGCUUUGUGC-CGGGCCUACAAUACAGUGACUGCCACCCACCUCCC-------ACCGCCCACAGCCCCUUG ....(((.((((.....((((((((....((((.(((((..(((.(((((((.....))-)))))))).....(((...)))...))).)).)))-------))))))))).))))))). ( -54.50) >DroSim_CAF1 33596 101 + 1 CGCGGAGUGGGCGGA-----------AAGUGGGCGGGGGACUAGUGCUUGGCUUUGUGC-CGGGCCUACAAUACAGUGACUGCCACCCACCUUUC-------GCCGCCCACAGAACCUUG ......((((((((.-----------..(((((.((.((..(((.(((((((.....))-))))))))((......)).)).)).))))).....-------.))))))))......... ( -45.90) >DroEre_CAF1 38763 108 + 1 CGCGGAGAGGGCGGA-----------AAGUGGGCGGAGGACUAGUGCUUGGCUUUGUGC-CGGGCCUACAAUACAGUGACUACCGCCCACCGCCCUCCGCCCUCCGCCCACCGCCCCUUG .((((...(((((((-----------....(((((((((..(((.(((((((.....))-)))))))).......(((.....))).......)))))))))))))))).))))...... ( -56.10) >DroWil_CAF1 123644 79 + 1 CGCGA-----GCACA---------------------AGGACUAGUGUUUGGCUUUGUGUUUGGGCCUAUAAUACAAA------UAUCCACCCUAC-------CCCAACCAU--CAUCUUC (.(((-----(((((---------------------(((.((((...))))))))))))))).).............------............-------.........--....... ( -15.00) >consensus CGCGGAGUGGGCGGA___________AAGUGGGCGGGGGACUAGUGCUUGGCUUUGUGC_CGGGCCUACAAUACAGUGACUGCCACCCACCUUCC_______CCCGCCCACAGCCCCUUG ...((.((((((((..............(((((.((.((..(((.(((((((.....)).))))))))((......)).)).)).))))).............))))))))....))... (-23.02 = -26.19 + 3.17)

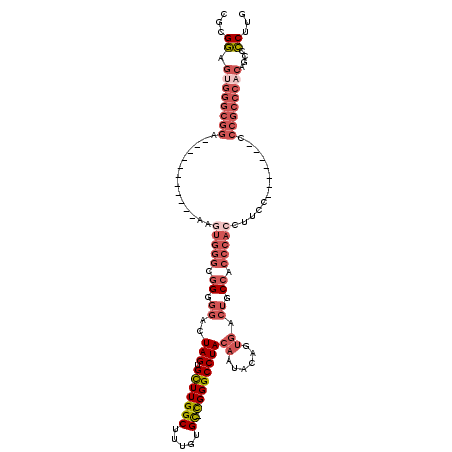
| Location | 1,437,046 – 1,437,147 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -24.71 |
| Energy contribution | -27.68 |
| Covariance contribution | 2.97 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1437046 101 - 22224390 CAAGGGGCUGUGGGCGGG-------GGAAGGUGGGUGGCAGUCACUGUAUUGUAGGCCCA-GCACAAAGCCAAGCACUAGUCCCCCGCCCAUUU-----------UCCGCCCACUCCGCG ....((((.(((((((((-------(((.(.(((((.(((((......)))))..)))))-.).....((...)).....))))))))))))..-----------...))))........ ( -47.70) >DroSec_CAF1 41766 112 - 1 CAAGGGGCUGUGGGCGGU-------GGGAGGUGGGUGGCAGUCACUGUAUUGUAGGCCCG-GCACAAAGCCAAGCACUAGUCCCCCGCCCACUUUCCGCCCACUUUCCGCCCACUCCGCG ...(.((..(((((((((-------(((((((((((((((((......))))(((((..(-((.....)))..)).))).....))))))))))))))))........)))))).)).). ( -52.60) >DroSim_CAF1 33596 101 - 1 CAAGGUUCUGUGGGCGGC-------GAAAGGUGGGUGGCAGUCACUGUAUUGUAGGCCCG-GCACAAAGCCAAGCACUAGUCCCCCGCCCACUU-----------UCCGCCCACUCCGCG ...((....((((((((.-------..(((((((((((((((......))))(((((..(-((.....)))..)).))).....))))))))))-----------))))))))).))... ( -44.80) >DroEre_CAF1 38763 108 - 1 CAAGGGGCGGUGGGCGGAGGGCGGAGGGCGGUGGGCGGUAGUCACUGUAUUGUAGGCCCG-GCACAAAGCCAAGCACUAGUCCUCCGCCCACUU-----------UCCGCCCUCUCCGCG ......((((.((((((((((((((((((((((...((..(((((......)).)))))(-((.....)))...)))).)))))))))))....-----------)))))))...)))). ( -60.10) >DroWil_CAF1 123644 79 - 1 GAAGAUG--AUGGUUGGG-------GUAGGGUGGAUA------UUUGUAUUAUAGGCCCAAACACAAAGCCAAACACUAGUCCU---------------------UGUGC-----UCGCG .......--.(((((..(-------((.((((..(((------.......)))..))))..)).)..)))))..(((.......---------------------.))).-----..... ( -14.80) >consensus CAAGGGGCUGUGGGCGGG_______GGAAGGUGGGUGGCAGUCACUGUAUUGUAGGCCCG_GCACAAAGCCAAGCACUAGUCCCCCGCCCACUU___________UCCGCCCACUCCGCG ...((....(((((((((..........((((((((((.....((((...(((.(((...........)))..))).))))...))))))))))...........))))))))).))... (-24.71 = -27.68 + 2.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:13 2006