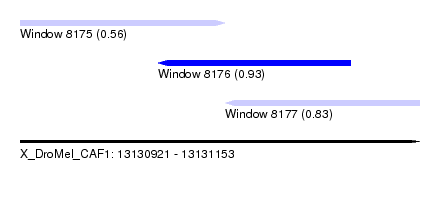
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,130,921 – 13,131,153 |
| Length | 232 |
| Max. P | 0.930479 |

| Location | 13,130,921 – 13,131,040 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -17.86 |
| Energy contribution | -20.87 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13130921 119 + 22224390 UACAAAUUAUUGAAAAUCACGACUUGUGGUGAACAGUCACGACCAAAGCAGUGACCUUAAGAAUUGCCCGCUCCAGCGUUAUCGAUAACACACCGAUAACGACUGCUAACGCAUACACA ....((((.((((...((((..(((((.((((....)))).))..)))..))))..)))).))))((......(((((((((((.........)))))))).))).....))....... ( -26.20) >DroSim_CAF1 2161 112 + 1 -------UAUAAAAAAUCGUGACUUGUGGUGAACAGUUACGACCGCAGCAGUGACCUCAAGAAUUACCCGCUCCAGCGUUAUCGAUAAACCGCCGAUAACGAGUGCUAACGCAUACACA -------.........((((((((.((.....))))))))))........(((..(((...........((....))(((((((.........))))))))))(((....)))..))). ( -28.70) >DroEre_CAF1 21519 115 + 1 AA---A-AAUGUAAAAACGUGACUCCCGGUGAACGGUCACGACCGCAGAAGUGACCUUAAAAAUAGCCCGCACUAGCAUUAUCGAUAACGCACCGAUAACGACUGCUAACGCAUACACA ..---.-...........(((.....(((((...((((((..........)))))).............((....)).............)))))........(((....)))..))). ( -25.60) >consensus _A___A_UAUAAAAAAUCGUGACUUGUGGUGAACAGUCACGACCGCAGCAGUGACCUUAAGAAUUGCCCGCUCCAGCGUUAUCGAUAACACACCGAUAACGACUGCUAACGCAUACACA .................((((((.(((.....))))))))).........(((.......................((((((((.........))))))))..(((....)))..))). (-17.86 = -20.87 + 3.00)


| Location | 13,131,001 – 13,131,113 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -32.35 |
| Energy contribution | -31.72 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13131001 112 - 22224390 GUGUGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGAUGUGUAUGCGUUAGCAGUCGUUAUCGG-UGUGUUAUCGAU ...((((((......)))....((((((.(((((.((((((.......))))))....((((......)))).))))).)))))).)))......(((((-((...))))))) ( -32.20) >DroSim_CAF1 2234 112 - 1 GUGUGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUCGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGAUGUGUAUGCGUUAGCACUCGUUAUCGG-CGGUUUAUCGAU (.(((((((......)))....((((((.(((((.((((((.......))))))....((((......)))).))))).)))))).)))).)...(((((-.......))))) ( -36.00) >DroEre_CAF1 21595 112 - 1 GUGCGCGCGGGAAAUCGCGAAAAAUGCACGCACAACUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGAUGUGUAUGCGUUAGCAGUCGUUAUCGG-UGCGUUAUCGAU ..((((((.((.(((.((....((((((..((((.((((((.......))))))....((((......)))).))))..))))))....)).))).)).)-)))))....... ( -37.70) >DroYak_CAF1 8980 113 - 1 GUGCGCGCGGGAAAACGCGGAAAAUGCACGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGAUGUGUAUGCGUUAGCAGUCGUUAUCGGAAGUGUUAUCGAU .(((.((((......))))...((((((..((((.((((((.......))))))....((((......)))).))))..)))))).)))((((.((.(....)...)).)))) ( -35.20) >consensus GUGCGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGAUGUGUAUGCGUUAGCAGUCGUUAUCGG_UGUGUUAUCGAU .(((.((((......))))...((((((.(((((.((((((.......))))))....((((......)))).))))).)))))).)))......(((((........))))) (-32.35 = -31.72 + -0.62)


| Location | 13,131,040 – 13,131,153 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -30.54 |
| Energy contribution | -30.48 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13131040 113 - 22224390 UUACUAUUGGGCGCAUUAAUUUUACGGCUACCGAAGGAUUGUGUGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGA ......(((((((((((((((((.(((...))).)))))).....((((......))))...)))))........((((((.......)))))).....))))))........ ( -30.10) >DroSim_CAF1 2273 113 - 1 UUACUAUUGGGCGCAUUAAUUUCACGGCUGCCGAAGGAUUGUGUGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUCGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGA ......(((((((((((.....(((((((......)).)))))..((((......))))...)))))........((((((.......)))))).....))))))........ ( -31.60) >DroEre_CAF1 21634 113 - 1 UCACUAUUGGGCGCAUUAAUUUCACGGCUGCCGGAGGAUUGUGCGCGCGGGAAAUCGCGAAAAAUGCACGCACAACUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGA ((.(((((((((((...(((((..(((...)))..)))))((((((((((....))))).....)))))))....((((((.......)))))).....))))))..))).)) ( -36.60) >DroYak_CAF1 9020 113 - 1 UCACGAUUGGGCGCAUCAAUUUUACGGCUGCCGUAGGAUUGUGCGCGCGGGAAAACGCGGAAAAUGCACGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGA ...(((((((((((...((((((((((...))))))))))(((((((((......)))).....)))))))....((((((.......)))))).....)))).))))).... ( -38.40) >consensus UCACUAUUGGGCGCAUUAAUUUCACGGCUGCCGAAGGAUUGUGCGCGCGGGAAAACGCGAAAAAUGCAAGCACAAUUUGCAAUUUACAUGCGAGAAAAUGCCCAAAUUGGCGA ......(((((((((((((((((.(((...))).)))))))....((((......))))....))))........((((((.......)))))).....))))))........ (-30.54 = -30.48 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:21 2006