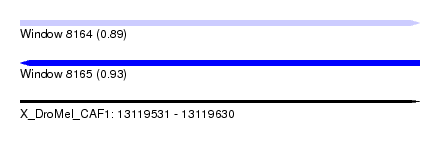
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,119,531 – 13,119,630 |
| Length | 99 |
| Max. P | 0.933434 |

| Location | 13,119,531 – 13,119,630 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -17.86 |
| Energy contribution | -17.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13119531 99 + 22224390 AAUCCACGACUAUUAGAGCUGGAACCACAUCGAUAUCUGUUCGAUACUAUCGAUACGGUCGUAGAAUUGGAAA-AUCGAUAAGUCUCUUGGAACAUCGAU ..((((.((((......(.((....)))((((((.....((((((.(((.((((...))))))).))))))..-)))))).))))...))))........ ( -20.50) >DroSec_CAF1 20731 99 + 1 AUUCCACGACUAUUAGAGCUGGAACCACAUCGAUAUCUAUUCGAUGCUAUCGAUGUGGUCGUAGGAUUGGAAA-UACGAUGAAUCUCUUGGAACAUCGAU .((((((........)..(((..((((((((((((.(........).))))))))))))..)))...))))).-..(((((..((....))..))))).. ( -29.40) >DroSim_CAF1 31826 98 + 1 AUUCCACGACUAUUAGAGCUGGAACCACAUCGAUAUCUAUUCGAUGCUAUCGAUGUGGUCGUAA-AUUGAAAA-UACGAUGAAUCUCUUGGAACAUCGAU ..((((.(.((.....)))))))((((((((((((.(........).)))))))))))).....-........-..(((((..((....))..))))).. ( -26.50) >DroYak_CAF1 20943 99 + 1 AUUCCACGACUAUUAGAGAUGGAACCACAUCGAUAUCUAUUCGGU-CUAUCGAUGUUUUUAAAGAGUUAGAAACAUCGAUGUUGUUCUUGACUAAUCGAU ......(((...((((....(((((.(((((((.......)))))-..((((((((((((((....)))))))))))))))).)))))...))))))).. ( -27.10) >consensus AUUCCACGACUAUUAGAGCUGGAACCACAUCGAUAUCUAUUCGAUGCUAUCGAUGUGGUCGUAGAAUUGGAAA_AACGAUGAAUCUCUUGGAACAUCGAU ..(((((..........).))))((((((((((((((.....))...)))))))))))).................((((...((.....))..)))).. (-17.86 = -17.93 + 0.06)



| Location | 13,119,531 – 13,119,630 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
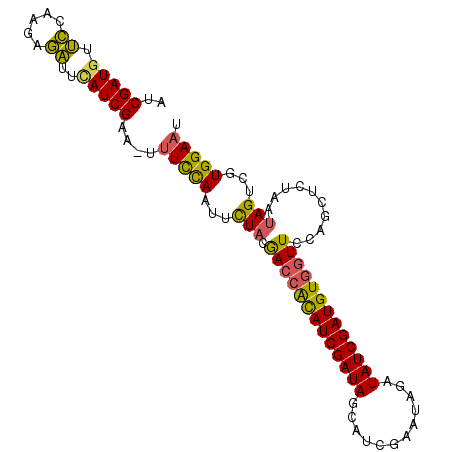
>X_DroMel_CAF1 13119531 99 - 22224390 AUCGAUGUUCCAAGAGACUUAUCGAU-UUUCCAAUUCUACGACCGUAUCGAUAGUAUCGAACAGAUAUCGAUGUGGUUCCAGCUCUAAUAGUCGUGGAUU (((((((.((.....))..)))))))-........((((((((((((((((((...((.....))))))))))))((....)).......)))))))).. ( -23.30) >DroSec_CAF1 20731 99 - 1 AUCGAUGUUCCAAGAGAUUCAUCGUA-UUUCCAAUCCUACGACCACAUCGAUAGCAUCGAAUAGAUAUCGAUGUGGUUCCAGCUCUAAUAGUCGUGGAAU ..(((((.((.....))..)))))..-.(((((...(((.(((((((((((((.(........).)))))))))))))..........)))...))))). ( -27.00) >DroSim_CAF1 31826 98 - 1 AUCGAUGUUCCAAGAGAUUCAUCGUA-UUUUCAAU-UUACGACCACAUCGAUAGCAUCGAAUAGAUAUCGAUGUGGUUCCAGCUCUAAUAGUCGUGGAAU ......((((((...((((..(((((-........-.)))))(((((((((((.(........).))))))))))).............)))).)))))) ( -27.80) >DroYak_CAF1 20943 99 - 1 AUCGAUUAGUCAAGAACAACAUCGAUGUUUCUAACUCUUUAAAAACAUCGAUAG-ACCGAAUAGAUAUCGAUGUGGUUCCAUCUCUAAUAGUCGUGGAAU ((((((..(((.........((((((((((.(((....))).))))))))))..-........)))))))))...(((((((..(.....)..))))))) ( -22.21) >consensus AUCGAUGUUCCAAGAGAUUCAUCGAA_UUUCCAAUUCUACGACCACAUCGAUAGCAUCGAAUAGAUAUCGAUGUGGUUCCAGCUCUAAUAGUCGUGGAAU ..(((((.((.....))..)))))....(((((...(((.(((((((((((((............)))))))))))))..........)))...))))). (-20.02 = -19.28 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:09 2006