| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,115,196 – 13,115,305 |
| Length | 109 |
| Max. P | 0.841633 |

| Location | 13,115,196 – 13,115,305 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13115196 109 + 22224390 -----UCCCACUGUACAGACCCACCAACC---CACCCAUUAUACCCCCCCACUUUUCGGCCCAAUGAGAGGAGCAACAAACGCACAGUGGUUCAAGCUGUGCAAAUAUUUAACUGAU -----......(((((((....((((...---...................(((((((......))))))).((.......))....)))).....))))))).............. ( -19.80) >DroSec_CAF1 16445 107 + 1 -----UCCCACUGUACAGAGCCACCAC-----CUCCCAUUAAACCCCCCCACUUUUCGGCCGAAUGAGAGGAGCAACAAACGCACAGUGGUUCAAGCUGUGCAAAUAUUUAACUGAU -----......(((((((((((((..(-----(((.((((...((............))...)))).)))).((.......))...)))))))....)))))).............. ( -25.70) >DroSim_CAF1 26855 107 + 1 -----UCCCACUGUACAGAGCCACCAC-----CCCCCAUUAAACCCCCCCACUUUUCGGCCGAAUGAGAGGAGCAACAAACGCACAGUGGUUCAAGCUGUGCAAAUAUUUAACUGAU -----......(((((((....(((((-----...................(((((((......))))))).((.......))...))))).....))))))).............. ( -22.50) >DroYak_CAF1 16615 111 + 1 -----UCCCACUGUACAGAGCCACCACCACGCCCCCCGGUGAAUCC-CCCUCUGUUCGGGCGAACGAGAGGAGCAACAAACGCACAGUGGUUCAAGCUGUGCAAAUAUUUAACUGAU -----......(((((((....(((((..((((....)))).....-.(((((((((....)))).))))).((.......))...))))).....))))))).............. ( -32.20) >DroAna_CAF1 22987 103 + 1 UAUUGUCCCACUGUA----GCCCCCCC------UACC----ACCCUACCCACCUCUAGACCAUGAAGCAGAAGCAACAAACGCACAGUGGGACAAGCUGUGUCAAUAUUUGAUUGAU ..((((((((((((.----((......------....----...(((........)))....((..((....))..))...)))))))))))))).....((((((.....)))))) ( -27.20) >consensus _____UCCCACUGUACAGAGCCACCAC_____CCCCCAUUAAACCCCCCCACUUUUCGGCCGAAUGAGAGGAGCAACAAACGCACAGUGGUUCAAGCUGUGCAAAUAUUUAACUGAU .......(((((((.....................................(((((((......))))))).((.......)))))))))........................... (-14.96 = -15.52 + 0.56)

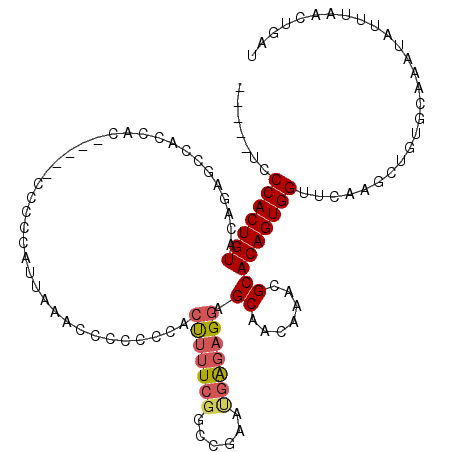
| Location | 13,115,196 – 13,115,305 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.74 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13115196 109 - 22224390 AUCAGUUAAAUAUUUGCACAGCUUGAACCACUGUGCGUUUGUUGCUCCUCUCAUUGGGCCGAAAAGUGGGGGGGUAUAAUGGGUG---GGUUGGUGGGUCUGUACAGUGGGA----- .((((((............))).))).((((((((((((..(((..((.(((((((.(((............))).))))))).)---)..)))..))..))))))))))..----- ( -31.40) >DroSec_CAF1 16445 107 - 1 AUCAGUUAAAUAUUUGCACAGCUUGAACCACUGUGCGUUUGUUGCUCCUCUCAUUCGGCCGAAAAGUGGGGGGGUUUAAUGGGAG-----GUGGUGGCUCUGUACAGUGGGA----- .((((((............))).))).((((((((((...((..(((((((((((..(((............)))..))))))))-----).))..))..))))))))))..----- ( -40.20) >DroSim_CAF1 26855 107 - 1 AUCAGUUAAAUAUUUGCACAGCUUGAACCACUGUGCGUUUGUUGCUCCUCUCAUUCGGCCGAAAAGUGGGGGGGUUUAAUGGGGG-----GUGGUGGCUCUGUACAGUGGGA----- .((((((............))).))).((((((((((...((..(((((((((((..(((............)))..))))))))-----).))..))..))))))))))..----- ( -39.30) >DroYak_CAF1 16615 111 - 1 AUCAGUUAAAUAUUUGCACAGCUUGAACCACUGUGCGUUUGUUGCUCCUCUCGUUCGCCCGAACAGAGGG-GGAUUCACCGGGGGGCGUGGUGGUGGCUCUGUACAGUGGGA----- .((((((............))).))).((((((((((...((..((((((((((((....)))).)))))-))...(((((.(...).))))))..))..))))))))))..----- ( -41.00) >DroAna_CAF1 22987 103 - 1 AUCAAUCAAAUAUUGACACAGCUUGUCCCACUGUGCGUUUGUUGCUUCUGCUUCAUGGUCUAGAGGUGGGUAGGGU----GGUA------GGGGGGGC----UACAGUGGGACAAUA .(((((.....)))))......((((((((((((((.(((.(((((((..((((........))))..))......----))))------).))).))----.)))))))))))).. ( -38.10) >consensus AUCAGUUAAAUAUUUGCACAGCUUGAACCACUGUGCGUUUGUUGCUCCUCUCAUUCGGCCGAAAAGUGGGGGGGUUUAAUGGGAG_____GUGGUGGCUCUGUACAGUGGGA_____ ...........................((((((((((...((..((.((((((((.((((............)))).)))))))).......))..))..))))))))))....... (-23.30 = -24.74 + 1.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:05 2006