| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,100,751 – 13,100,865 |
| Length | 114 |
| Max. P | 0.634072 |

| Location | 13,100,751 – 13,100,865 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -42.28 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
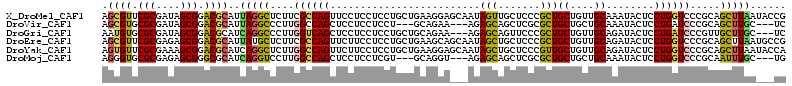
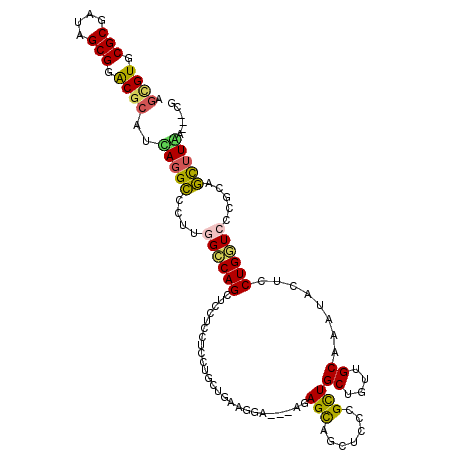
>X_DroMel_CAF1 13100751 114 + 22224390 AGCGUUCGCGAUAGCGGACGCAUUAGGCUCUUCGCCAGUUCCUCCUCCUGCUGAAGGAGCAAUAGUUGCUCCCGCUGCUGUUGCAAAUACUCCUGGUCCCGCAGCUUAAUACCG .((((((((....))))))))(((((((((...(((((.......((((.....))))((((((((.((....)).))))))))........)))))...).)))))))).... ( -46.70) >DroVir_CAF1 7374 105 + 1 AGCGUGCGCGAUAGCGGACGCAUUAGGCCCUUGGCCAGCUCCUCCUCCU---GCAGAA---AGAGCAGCUCGCGCUGCUGCUGCAAAUACUCCUGAUCCCGCAGCUUGC---UC .((((.(((....))).))))....((((...)))).............---((((..---..((((((....))))))(((((................)))))))))---.. ( -38.79) >DroGri_CAF1 1903 108 + 1 AAUGUGCGCGAUAGCGGACGCAUCAGGCCCUUGGUCAGCUCCUCCUCCUGCUGCAGAA---AGAGCAGUUCCCGCUGCUGUUGCAGAUACUCCUGAUCCCGUUGCUUGC---UC .......((((((((((....((((((.....((......))........((((((..---..((((((....)))))).)))))).....)))))).)))))).))))---.. ( -33.20) >DroEre_CAF1 1842 114 + 1 AGCGUUCGCGAGAGCGGACGCAUUAUGCUCUUCGCCAGUUCUUCCUCCUGCUGAAGCAGCAAUAGCUGCUCCCGCUGCUGUUGCAGAUACUCCUGGUCCCGCAGCUUAAUGCCG ...((((((....))))))((((((.((((...(((((..........(((....)))((((((((.((....)).))))))))........)))))...).))).)))))).. ( -44.90) >DroYak_CAF1 1794 114 + 1 AGUGUUCGCGAAAGCGGACGCAUCAGGCUCUUGGCCAGUUCUUCCUCCUGCUGAAGGAGCAAUAGCUGCUCCCGUUGCUGUUGCAGAUACUCCUGGUCCCGCAGCUUAAUACCA .((((((((....))))))))...((((((..((((((.......((((.....))))((((((((.((....)).))))))))........))))))..).)))))....... ( -47.30) >DroMoj_CAF1 3857 105 + 1 AGGGUGCGCGAGAGCGGGCGCAUCAGGUCCUUGGCCAGCUCCUCCUCGU---GCAGGU---AGAGCAGCUCGCGCUGCUGCUGCAAAUACUCCUGGUCCCGCAAUUUGC---UG .(((((((((((((.((((......(((.....))).)))))).)))))---))).((---((((((((....)))))).)))).............)))((.....))---.. ( -42.80) >consensus AGCGUGCGCGAUAGCGGACGCAUCAGGCCCUUGGCCAGCUCCUCCUCCUGCUGAAGGA___AGAGCAGCUCCCGCUGCUGUUGCAAAUACUCCUGGUCCCGCAGCUUAA___CG .((((.(((....))).))))..(((((....((((((.........................(((.......)))((....))........)))))).....)))))...... (-22.93 = -22.55 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:00 2006