
| Sequence ID | X_DroMel_CAF1 |
|---|---|
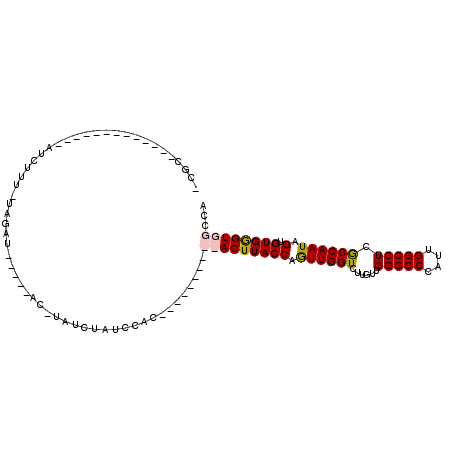
| Location | 13,090,496 – 13,090,602 |
| Length | 106 |
| Max. P | 0.999577 |

| Location | 13,090,496 – 13,090,602 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.57 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13090496 106 + 22224390 -CGCAUACUUUGGACGCAUCUUU-CAGAU-----ACUUAUCUAUCCACUUAACUAUCCACUUACCAGUUGGUCUUGUUGGGCCCAUUGGCCUCGCCAAUAGUGUGGGUGGCCA -.........(((..........-.((((-----....))))..............(((((((((.((((((......(((((....))))).)))))).).))))))))))) ( -26.50) >DroSec_CAF1 41010 82 + 1 -CGC-------------AUCUUU-UAGAU-----AC-UAUCUAUCCAC----------ACUUACCAGUUGGUCUUGUUGGGCCCAUUGGCCUCGCCAAUAGUGUGGGUGGCUA -...-------------(((...-..)))-----..-...((((((((----------(((.....((((((......(((((....))))).)))))))))))))))))... ( -27.00) >DroSim_CAF1 34275 82 + 1 -CGC-------------AUCUUU-UAGAU-----AC-UAUCUAUCCAC----------ACUUACCAGUUGGUCUUGUUGGGCCCAUUGGCCUCGCCAAUAGUGUGGGUGGCUA -...-------------(((...-..)))-----..-...((((((((----------(((.....((((((......(((((....))))).)))))))))))))))))... ( -27.00) >DroYak_CAF1 47757 110 + 1 AUGUAUAUUUUGGAUCCUUCUUCGAUGAUCAAUGAUAUAUGUAUGAAUG---UUAGCCACUUACCAAUUGGUCUUGUUGGGCCCAUUGGCCUCACCAAUCGUGUGAGUGGCCA (((((((((...((((.((....)).))))...))))))))).......---...((((((((((.((((((......(((((....))))).)))))).).))))))))).. ( -36.30) >consensus _CGC_____________AUCUUU_UAGAU_____AC_UAUCUAUCCAC__________ACUUACCAGUUGGUCUUGUUGGGCCCAUUGGCCUCGCCAAUAGUGUGGGUGGCCA ........................................................(((((((((.((((((......(((((....))))).)))))).).))))))))... (-20.14 = -20.57 + 0.44)

| Location | 13,090,496 – 13,090,602 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.57 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13090496 106 - 22224390 UGGCCACCCACACUAUUGGCGAGGCCAAUGGGCCCAACAAGACCAACUGGUAAGUGGAUAGUUAAGUGGAUAGAUAAGU-----AUCUG-AAAGAUGCGUCCAAAGUAUGCG- (((.(..((((((((((.(((.((((....)))))......(((....)))..)).))))))...))))........((-----(((..-...)))))).))).........- ( -30.80) >DroSec_CAF1 41010 82 - 1 UAGCCACCCACACUAUUGGCGAGGCCAAUGGGCCCAACAAGACCAACUGGUAAGU----------GUGGAUAGAUA-GU-----AUCUA-AAAGAU-------------GCG- .......(((((((.(((..(.((((....)))))..))).(((....))).)))----------)))).......-((-----(((..-...)))-------------)).- ( -27.40) >DroSim_CAF1 34275 82 - 1 UAGCCACCCACACUAUUGGCGAGGCCAAUGGGCCCAACAAGACCAACUGGUAAGU----------GUGGAUAGAUA-GU-----AUCUA-AAAGAU-------------GCG- .......(((((((.(((..(.((((....)))))..))).(((....))).)))----------)))).......-((-----(((..-...)))-------------)).- ( -27.40) >DroYak_CAF1 47757 110 - 1 UGGCCACUCACACGAUUGGUGAGGCCAAUGGGCCCAACAAGACCAAUUGGUAAGUGGCUAA---CAUUCAUACAUAUAUCAUUGAUCAUCGAAGAAGGAUCCAAAAUAUACAU ((((((((.((.(((((((((.((((....)))))......)))))))))).)))))))).---..........(((((....((((..........))))....)))))... ( -32.10) >consensus UAGCCACCCACACUAUUGGCGAGGCCAAUGGGCCCAACAAGACCAACUGGUAAGU__________GUGGAUAGAUA_GU_____AUCUA_AAAGAU_____________GCG_ ............(((((.(((.((((....)))))......(((....)))..............)).)))))........................................ (-13.80 = -14.18 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:59 2006