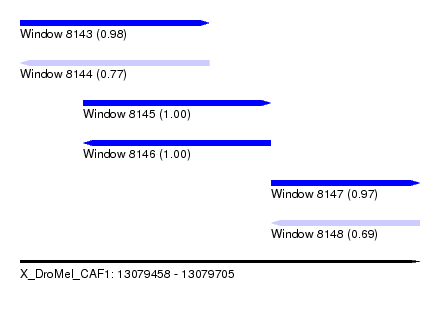
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,079,458 – 13,079,705 |
| Length | 247 |
| Max. P | 0.999876 |

| Location | 13,079,458 – 13,079,575 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -16.09 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079458 117 + 22224390 GUGAGCAAAGUUAUUUCGAAUUGAGU-UUGCGUCAAGGUGAAGAUCAGCGCUGAGUAAGAUAGGUGCAAAUAUCUAGUAA--AAUUACUCAGCGCUGGAAUAAAACAUAUAUAUAUGUGC .((((((((.(((........))).)-)))).))).........((((((((((((((..((((((....))))))....--..))))))))))))))......(((((....))))).. ( -37.60) >DroSec_CAF1 28633 110 + 1 GUGAGUAAAGUUAUACAGAAUUGAGU-UUGCGUCAAGGUGAAGAUCAGCGCUGAGUAAGAUAUGUGCAAAUAUCUAGUAA--AAUUACUCAGCGUCGGAAUAAAACAUAUAUG------- .((((((((.(((........))).)-)))).)))..((.....((.((((((((((((((((......)))))).....--...))))))))))..)).....)).......------- ( -26.70) >DroEre_CAF1 28641 114 + 1 GCGAGUAAAGUUAUUCAGAAUUGCGA-UUGCGUCCAGAGGAUGAUCCGUGGUGAUUAAG-----AGCAAAUAUCUAGAAAAGAAUUAAUCAGCGCCGGAAUAAAACAUAUAUACUCGUAU (((((((...................-...(((((...))))).((((((.(((((((.-----..(..............)..))))))).)).))))............))))))).. ( -25.54) >DroYak_CAF1 31219 118 + 1 GUGAGUAAAGUUAUUCAGAAUUGAGUUUUGCAUCAAUGUGAAUAUCAUUGCCGAGUAAGCU-GGUGCAAAUAUCUAGCAAA-AAUUACUCAGCAGCAAAAUAAAACCUACAUAUAUAUGU (((.((...(.((((((..(((((........))))).)))))).).((((.(((((((((-((.........)))))...-..)))))).)))).........)).))).......... ( -24.40) >consensus GUGAGUAAAGUUAUUCAGAAUUGAGU_UUGCGUCAAGGUGAAGAUCAGCGCUGAGUAAGAU_GGUGCAAAUAUCUAGUAA__AAUUACUCAGCGCCGGAAUAAAACAUAUAUAUAUGUGU ((((....((((......)))).....)))).............((.(((((((((((......(((.........))).....))))))))))).))...................... (-16.09 = -15.53 + -0.56)

| Location | 13,079,458 – 13,079,575 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079458 117 - 22224390 GCACAUAUAUAUAUGUUUUAUUCCAGCGCUGAGUAAUU--UUACUAGAUAUUUGCACCUAUCUUACUCAGCGCUGAUCUUCACCUUGACGCAA-ACUCAAUUCGAAAUAACUUUGCUCAC ..(((((....))))).......((((((((((((...--.....(((((........)))))))))))))))))..........(((.((((-(................)))))))). ( -27.69) >DroSec_CAF1 28633 110 - 1 -------CAUAUAUGUUUUAUUCCGACGCUGAGUAAUU--UUACUAGAUAUUUGCACAUAUCUUACUCAGCGCUGAUCUUCACCUUGACGCAA-ACUCAAUUCUGUAUAACUUUACUCAC -------..(((((.......((.(.(((((((((...--.....((((((......)))))))))))))))).))........((((.....-..))))....)))))........... ( -18.30) >DroEre_CAF1 28641 114 - 1 AUACGAGUAUAUAUGUUUUAUUCCGGCGCUGAUUAAUUCUUUUCUAGAUAUUUGCU-----CUUAAUCACCACGGAUCAUCCUCUGGACGCAA-UCGCAAUUCUGAAUAACUUUACUCGC ...((((((....(((((...(((((.(.(((((((.(((.....)))........-----.))))))).)..((.....)).))))).((..-..))......)))))....)))))). ( -19.10) >DroYak_CAF1 31219 118 - 1 ACAUAUAUAUGUAGGUUUUAUUUUGCUGCUGAGUAAUU-UUUGCUAGAUAUUUGCACC-AGCUUACUCGGCAAUGAUAUUCACAUUGAUGCAAAACUCAAUUCUGAAUAACUUUACUCAC .(((......(((((......)))))((((((((((((-..(((.........)))..-)).))))))))))))).((((((.(((((........)))))..))))))........... ( -23.50) >consensus ACACAUAUAUAUAUGUUUUAUUCCGGCGCUGAGUAAUU__UUACUAGAUAUUUGCACC_AUCUUACUCAGCACUGAUCUUCACCUUGACGCAA_ACUCAAUUCUGAAUAACUUUACUCAC .......................(((((((((((((..........................)))))))))))))............................................. (-12.10 = -12.22 + 0.12)


| Location | 13,079,497 – 13,079,613 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.54 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079497 116 + 22224390 AAGAUCAGCGCUGAGUAAGAUAGGUGCAAAUAUCUAGUAA--AAUUACUCAGCGCUGGAAUAAAACAUAUAUAUAUGUGCAUACAUACA-AUAUAUGCAUAAAACGCAAAAAUCCCAAU- ....((((((((((((((..((((((....))))))....--..)))))))))))))).................((((((((......-...))))))))..................- ( -33.60) >DroSec_CAF1 28672 91 + 1 AAGAUCAGCGCUGAGUAAGAUAUGUGCAAAUAUCUAGUAA--AAUUACUCAGCGUCGGAAUAAAACAUAUAUG--------------------------UAAACCACGGAAAUCCCAGU- ....((.((((((((((((((((......)))))).....--...)))))))))).))...............--------------------------........((.....))...- ( -21.00) >DroEre_CAF1 28680 95 + 1 AUGAUCCGUGGUGAUUAAG-----AGCAAAUAUCUAGAAAAGAAUUAAUCAGCGCCGGAAUAAAACAUAUAUACUCGUAUCUACAUGAACGUAUAUGC------CA-------------- ....((((((.(((((((.-----..(..............)..))))))).)).))))......(((((((..((((......))))..))))))).------..-------------- ( -19.04) >DroYak_CAF1 31259 118 + 1 AAUAUCAUUGCCGAGUAAGCU-GGUGCAAAUAUCUAGCAAA-AAUUACUCAGCAGCAAAAUAAAACCUACAUAUAUAUGUAUAUACAGACAUAUAUGCAUAAGCCACAGAAAUCGCAGUG .....((((((.(((((((((-((.........)))))...-..)))))).........................((((((((((......)))))))))).............)))))) ( -24.40) >consensus AAGAUCAGCGCUGAGUAAGAU_GGUGCAAAUAUCUAGUAA__AAUUACUCAGCGCCGGAAUAAAACAUAUAUAUAUGUGUAUACAUA_A_AUAUAUGC_UAAACCACAGAAAUCCCAGU_ ....((.(((((((((((......(((.........))).....))))))))))).)).............................................................. (-14.62 = -14.25 + -0.38)


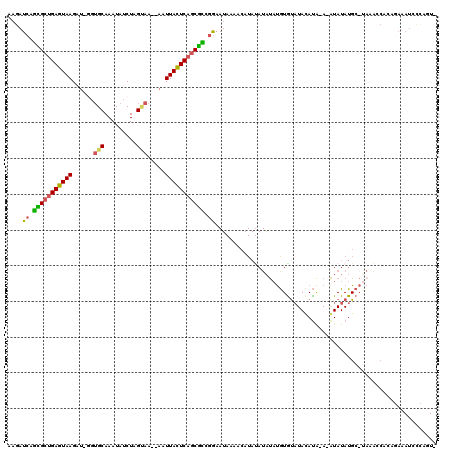
| Location | 13,079,497 – 13,079,613 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.54 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079497 116 - 22224390 -AUUGGGAUUUUUGCGUUUUAUGCAUAUAU-UGUAUGUAUGCACAUAUAUAUAUGUUUUAUUCCAGCGCUGAGUAAUU--UUACUAGAUAUUUGCACCUAUCUUACUCAGCGCUGAUCUU -...(((((.............((((((((-.((((((....)))))))))))))).......((((((((((((...--.....(((((........)))))))))))))))))))))) ( -34.40) >DroSec_CAF1 28672 91 - 1 -ACUGGGAUUUCCGUGGUUUA--------------------------CAUAUAUGUUUUAUUCCGACGCUGAGUAAUU--UUACUAGAUAUUUGCACAUAUCUUACUCAGCGCUGAUCUU -...((((((..((..((..(--------------------------((....)))...))..)).(((((((((...--.....((((((......)))))))))))))))..)))))) ( -21.00) >DroEre_CAF1 28680 95 - 1 --------------UG------GCAUAUACGUUCAUGUAGAUACGAGUAUAUAUGUUUUAUUCCGGCGCUGAUUAAUUCUUUUCUAGAUAUUUGCU-----CUUAAUCACCACGGAUCAU --------------.(------(((((((..(((..........)))..))))))))....((((..(.(((((((.(((.....)))........-----.))))))).).)))).... ( -16.50) >DroYak_CAF1 31259 118 - 1 CACUGCGAUUUCUGUGGCUUAUGCAUAUAUGUCUGUAUAUACAUAUAUAUGUAGGUUUUAUUUUGCUGCUGAGUAAUU-UUUGCUAGAUAUUUGCACC-AGCUUACUCGGCAAUGAUAUU ....((((.......(((((((..((((((((........))))))))..))))))).....))))((((((((((((-..(((.........)))..-)).))))))))))........ ( -30.50) >consensus _ACUGGGAUUUCUGUGGUUUA_GCAUAUAU_U_UAUGUAUACACAUAUAUAUAUGUUUUAUUCCGGCGCUGAGUAAUU__UUACUAGAUAUUUGCACC_AUCUUACUCAGCACUGAUCUU ...............................................................(((((((((((((..........................)))))))))))))..... (-12.10 = -12.22 + 0.12)



| Location | 13,079,613 – 13,079,705 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -30.43 |
| Energy contribution | -30.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079613 92 + 22224390 -------GAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUCACAGCUCCCCAGCACUUUAGCUCCACGAUUUC -------(((((((((((((((.(((........(.(((....((((...))))..))))(.....)...)))..)))))))))))))))......... ( -34.40) >DroSec_CAF1 28763 92 + 1 -------GAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUUUUAGCUCCGCAGCACUUUAGCUCCACGAUUUC -------(((((((((((((((.((((((..(..(.(((....((((...))))..))))..)...))).))).).))))))))))))))......... ( -33.40) >DroYak_CAF1 31377 99 + 1 AGUGAGUGAGUUGAAGUGCUGGGAGCAAAUCGAAGAGGUUUUUCUGGUCCCCAGUUGCCCGACACUCGUAGCUCCCCAGCACUUAAGCUCCCCGCUUUC ((((.(.(((((.(((((((((((((....(((.(.(((....((((...))))..)))).....)))..))).)))))))))).)))))).))))... ( -40.00) >consensus _______GAGUUGAAGUGCUGGAAGCAAAUGGAAGAGGUUUUUCUGGUCUCCAGUUGCCCGACACUCAUAGCUCCCCAGCACUUUAGCUCCACGAUUUC .......(((((((((((((((.(((.....(..(.(((....((((...))))..))))..).......))).).))))))))))))))......... (-30.43 = -30.77 + 0.33)


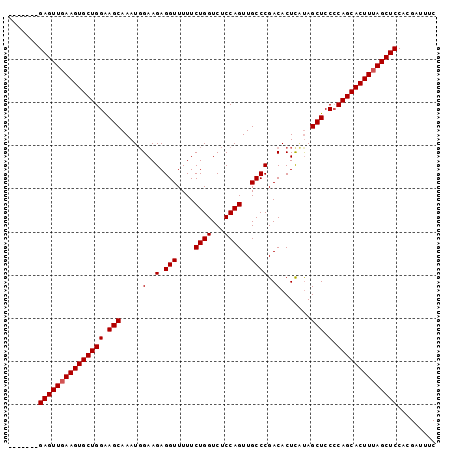
| Location | 13,079,613 – 13,079,705 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.93 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -27.57 |
| Energy contribution | -27.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13079613 92 - 22224390 GAAAUCGUGGAGCUAAAGUGCUGGGGAGCUGUGAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUC------- .........(((...((((((((((.(((.(((.....(((....)))(((...........)))...)))..)))))))))))))...)))------- ( -27.60) >DroSec_CAF1 28763 92 - 1 GAAAUCGUGGAGCUAAAGUGCUGCGGAGCUAAAAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUC------- .........(((...((((((((.(((((...(((((.(((....)))(((...........)))...))))))))))))))))))...)))------- ( -26.90) >DroYak_CAF1 31377 99 - 1 GAAAGCGGGGAGCUUAAGUGCUGGGGAGCUACGAGUGUCGGGCAACUGGGGACCAGAAAAACCUCUUCGAUUUGCUCCCAGCACUUCAACUCACUCACU ...((.((((((...((((((((((.(((..((((....((....))((((..........))))))))....)))))))))))))...))).))).)) ( -36.10) >consensus GAAAUCGUGGAGCUAAAGUGCUGGGGAGCUAAGAGUGUCGGGCAACUGGAGACCAGAAAAACCUCUUCCAUUUGCUUCCAGCACUUCAACUC_______ .........(((...((((((((.(((((.........(((....)))(((...........)))........)))))))))))))...)))....... (-27.57 = -27.13 + -0.44)


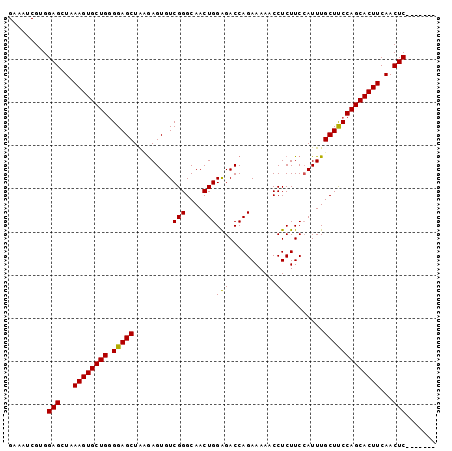
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:54 2006