
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,065,326 – 13,065,431 |
| Length | 105 |
| Max. P | 0.917905 |

| Location | 13,065,326 – 13,065,431 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.34 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -11.69 |
| Energy contribution | -10.83 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
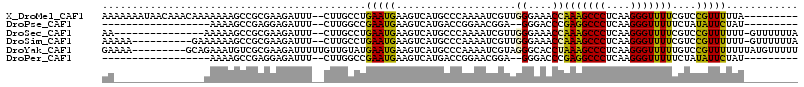
>X_DroMel_CAF1 13065326 105 + 22224390 AAAAAAAUAACAAACAAAAAAAGCCGCGAAGAUUU--CUUGCCUGAAUGAAGUCAUGCCCAAAAUCGUUGGGAAACCAAAGCCCUCAAGGGUUUUCGUCCGUUUUUA--------- ..................((((((.((((((((((--...((.(((......))).))...))))).((((....))))(((((....))))))))).).)))))).--------- ( -25.20) >DroPse_CAF1 17979 85 + 1 ------------------AAAAGCCGAGGAGAUUU--CUUGGCCGAAUGAAGUCAUGACCGGAACGGA--GGGACCCGAGGCCCUCAAGGGUUUUUCUAUAUUCUAU--------- ------------------....((((((((...))--)))))).(((((.((....((((......((--(((.((...)))))))...))))...)).)))))...--------- ( -26.70) >DroSec_CAF1 15659 98 + 1 AA---------------AAAAAGCCGCGAAGAUUU--CUUGCCUGAAUGAAGUCAUGCCCAAAAUCGUUGGGAAACCAAAGCCCUCAAGGGUUUUCGUCCGUUUUUU-GUUUUUUA ..---------------(((((((.((((((((((--...((.(((......))).))...))))).((((....))))(((((....))))))))).).)))))))-........ ( -26.10) >DroSim_CAF1 16854 103 + 1 AAAAA----------GAAAAAAGCCGCGAAGAUUU--CUUGCCUGAAUGAAGUCAUGCCCAAAAUCGUUGGGAAACCAAAGCCCUCAAGGGUUUUCGUCCGUUUUUU-GUUUUUUA (((((----------(.(((((((.((((((((((--...((.(((......))).))...))))).((((....))))(((((....))))))))).).)))))))-.)))))). ( -28.30) >DroYak_CAF1 15897 107 + 1 GAAAA---------GCAGAAAUGUCGCGAAGAUUUUUGUUGUAUGAAUGAAGUCAUGCCCAAAAUCGUAGGGCACCUAAAGCCCUCAAGGGUUUUUGUCCGUUUUUUUAUGUUUUU .((((---------(((((((((..(((((((((((((..((((((......))))))..........(((((.......)))))))))))))))))).))))))....))))))) ( -28.80) >DroPer_CAF1 17195 85 + 1 ------------------AAAAGCCGAGGAGAUUU--CUUGGCCGAAUGAAGUCAUGACCGGAACGGA--GGGACCCGAGGCCCUCAAGGGUUUUUCUAUAUUCUAU--------- ------------------....((((((((...))--)))))).(((((.((....((((......((--(((.((...)))))))...))))...)).)))))...--------- ( -26.70) >consensus AA_______________AAAAAGCCGCGAAGAUUU__CUUGCCUGAAUGAAGUCAUGCCCAAAAUCGUUGGGAAACCAAAGCCCUCAAGGGUUUUCGUCCGUUUUUU_________ ............................................(((((....................((....))(((((((....)))))))....)))))............ (-11.69 = -10.83 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:47 2006