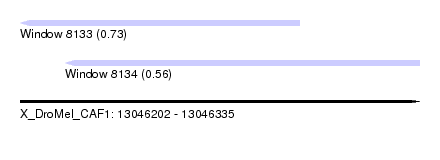
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,046,202 – 13,046,335 |
| Length | 133 |
| Max. P | 0.734810 |

| Location | 13,046,202 – 13,046,295 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -10.26 |
| Energy contribution | -11.90 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13046202 93 - 22224390 GUGGUGCUAGAGGUGGAAGAAAAAGAAUAAUCGAUGCUUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUUAAUACCCCACAAGUAUC ..((((((.(.((.((..............((((((((....-...))))))))((((((((((...))))))))))....)))).).)))))) ( -28.70) >DroSec_CAF1 15628 93 - 1 GUGGUGCUAGAGGUGGAGAAAAAAGUCUAAUCGAUACCUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUGAAUACCGCACGCGGAUC (((((((((.(((((..((...........))..)))))...-..))))......(((((((((...)))))))))....)))))......... ( -24.10) >DroSim_CAF1 15168 93 - 1 GUGGUGCUAGAGGUGGAGAAAAAAGACUAAUCGAUACCUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUGAAUACCGCACGAGGAUC (((((((((.(((((..((...........))..)))))...-..))))......(((((((((...)))))))))....)))))......... ( -24.10) >DroEre_CAF1 16333 91 - 1 GUGGUGUUAGAGGUGGUGGAAAAAAAAUAAUCGAUAUUUAUCCGCUGCUAUCGCAAU-UGCAGAAAUUUCUUAAGAAAAUACCAGA--AAUUGC .(((((((.((((..(((((....(((((.....))))).)))))..)).))((...-.))................)))))))..--...... ( -20.30) >DroYak_CAF1 16735 89 - 1 GUGGUGCUAGAGGUGGGGAAAAAUUAAUAAUCGAUGUUUAUGCG---CUAUCGCAAUUUGCUGAAAUUUCGCAAGAAAAUACCAGA--AAUUCU (((((((((((.((.((.............)).)).)))).)))---)))).(.(((((.(((...((((....))))....))))--))))). ( -18.42) >consensus GUGGUGCUAGAGGUGGAGAAAAAAGAAUAAUCGAUACUUAUC_GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUAAAUACCACAC_AGUAUC ((((((........................(((((((..........)))))))..((((((((...))))))))....))))))......... (-10.26 = -11.90 + 1.64)



| Location | 13,046,217 – 13,046,335 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13046217 118 - 22224390 ACUUACCGACAAAUUUUGAUGCCCAAGCAACAACACGCAUGUGGUGCUAGAGGUGGAAGAAAAAGAAUAAUCGAUGCUUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUUAA .(((.(((.....(((((..(((((.((........)).)).)))..))))).))))))...........((((((((....-...))))))))((((((((((...)))))))))).. ( -31.30) >DroSec_CAF1 15643 118 - 1 ACUUACUGACAAAUUUUGAUGCCCAAGCAACAACACACAUGUGGUGCUAGAGGUGGAGAAAAAAGUCUAAUCGAUACCUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUGAA .....(((.((((((((((((((((((((....(((....))).)))).....))).............((((((....)))-))).))))))))))))))))................ ( -32.80) >DroSim_CAF1 15183 118 - 1 ACUUACCGACAAAUUUUGAUGCCCAAGCAACAACACACAUGUGGUGCUAGAGGUGGAGAAAAAAGACUAAUCGAUACCUAUC-GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUGAA ......((((...((((..(((((.((((....(((....))).)))).).))))..))))...(.(((.(((((....)))-))))))))))..(((((((((...)))))))))... ( -31.60) >DroEre_CAF1 16346 118 - 1 ACUUACCGACAAAUUUUGAUGCCCAAGCAACAACACACAUGUGGUGUUAGAGGUGGUGGAAAAAAAAUAAUCGAUAUUUAUCCGCUGCUAUCGCAAU-UGCAGAAAUUUCUUAAGAAAA .((((.....(((((((..(((....((((((.(((....))).)))).((((..(((((....(((((.....))))).)))))..)).))))...-.))))))))))..)))).... ( -23.50) >DroYak_CAF1 16748 116 - 1 ACUUACCGACAAUUUUUGAUGCCCAAGCAACAACACACAUGUGGUGCUAGAGGUGGGGAAAAAUUAAUAAUCGAUGUUUAUGCG---CUAUCGCAAUUUGCUGAAAUUUCGCAAGAAAA ......((.((((((((..(((((.((((....(((....))).)))).).))))..)))))..................((((---....))))..))).))...((((....)))). ( -22.10) >consensus ACUUACCGACAAAUUUUGAUGCCCAAGCAACAACACACAUGUGGUGCUAGAGGUGGAGAAAAAAGAAUAAUCGAUACUUAUC_GAUAGCGUCGAAGUUUGCAGAAAUUUUGCAAAUAAA .......(.(((((((((((((...((((....(((....))).))))..(((((..((...........))..)))))........)))))))))))))).................. (-18.56 = -18.12 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:42 2006