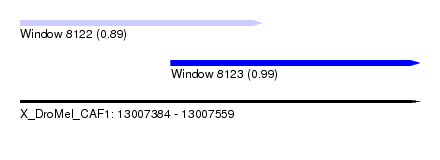
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 13,007,384 – 13,007,559 |
| Length | 175 |
| Max. P | 0.992734 |

| Location | 13,007,384 – 13,007,490 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.84 |
| Mean single sequence MFE | -14.23 |
| Consensus MFE | -9.33 |
| Energy contribution | -9.00 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13007384 106 + 22224390 GGCUAACUUAUGGCCAAAGGGAAUUCACCAUCGUAAGCAGUAAAAAUGAG-AAGAAAAAAAAUCAUAAAAGAAGAGCACAGAACAGAUUUUCAACGCGCUGGAACAA (((((.....)))))............(((.(((...........((((.-...........))))....(((((.(........).)))))...))).)))..... ( -15.30) >DroEre_CAF1 17880 91 + 1 GGCUAACUUAUAGCCAUAGGGAACCCA------------ACAAAAACCAAAUAAA--AUAAACCUCAAAAGAAUA--AGAGAACAGAUUUUCAACGCGUUGAAACAA (((((.....)))))...((....)).------------................--......(((.........--.))).......((((((....))))))... ( -13.20) >DroYak_CAF1 15209 97 + 1 GGCUAACUUAUGGCCAAAGGGAAUCCACAAACGUAAGCAGCAAAAAGCAU-UAAA--AAAA-----AAAUAUAUA--AGAGAACAGAUUUUCAACGCGUUGAAACAA (((((.....)))))...((....))...(((((.....((.....))..-....--....-----.........--.(((((....)))))...)))))....... ( -14.20) >consensus GGCUAACUUAUGGCCAAAGGGAAUCCAC_A_CGUAAGCAGCAAAAACCAA_UAAA__AAAAA_C__AAAAGAAUA__AGAGAACAGAUUUUCAACGCGUUGAAACAA (((((.....)))))...((....))..............................................................((((((....))))))... ( -9.33 = -9.00 + -0.33)

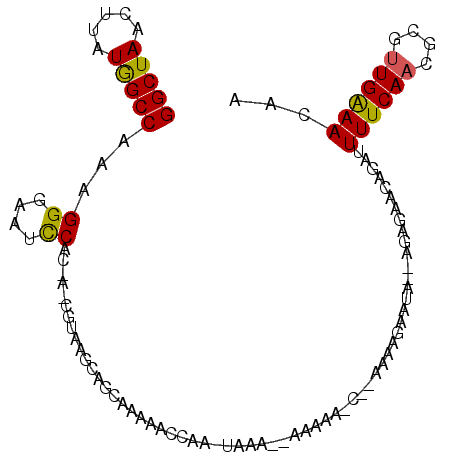

| Location | 13,007,450 – 13,007,559 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -22.22 |
| Energy contribution | -21.66 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 13007450 109 + 22224390 AAAGAAGAGCACAGAACAGAUUUUCAACGCGCUGGAACAAACAGAUGCUCUUGCCUAUUUGGCUCCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCA .............((((((((((((...((((((.......))).)))............((((((((...............)))))))))))).))))))))..... ( -26.46) >DroSec_CAF1 12733 109 + 1 AAAGAAGAGCACAAAACAGAUUUCCAACGCGCUGAAACAAACAGAUGCUCUUGCCUAUUUGGCUCCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCA .............((((((((.......((((((.......))).))).........(((((((((((...............)))))))))))..))))))))..... ( -23.76) >DroSim_CAF1 11647 109 + 1 AAAGAAGAGCACAGAACAGAUUUCCAACGCGCUGAAACAAACAGAUGCUCUUGCCUAUUUGGCUCCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCA .............((((((((.......((((((.......))).))).........(((((((((((...............)))))))))))..))))))))..... ( -24.16) >DroEre_CAF1 17933 107 + 1 AAAGAAUA--AGAGAACAGAUUUUCAACGCGUUGAAACAAACAGAUGCUCUUGCCUAUUUGGCUUCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCG ........--...((((((((((((...(((((..........)))))............((((((((...............)))))))))))).))))))))..... ( -21.26) >DroYak_CAF1 15268 107 + 1 AAUAUAUA--AGAGAACAGAUUUUCAACGCGUUGAAACAAACAGAUGCUGUUGCCUAUUUGGCUCCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCA ........--...((((((((...(((((((((..........))))).))))....(((((((((((...............)))))))))))..))))))))..... ( -25.86) >consensus AAAGAAGAGCACAGAACAGAUUUUCAACGCGCUGAAACAAACAGAUGCUCUUGCCUAUUUGGCUCCAUAAAUUUCUUAAGCAAAUGGGGCCGAAAUAUUUGUUUAACCA .............((((((((.......((((((.......))).))).........(((((((((((...............)))))))))))..))))))))..... (-22.22 = -21.66 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:32 2006