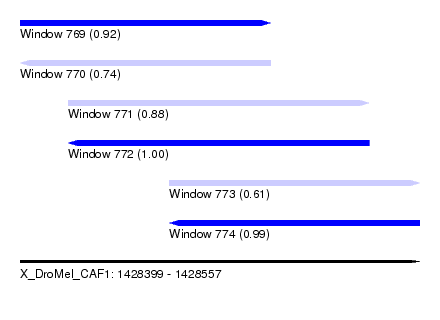
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,428,399 – 1,428,557 |
| Length | 158 |
| Max. P | 0.996318 |

| Location | 1,428,399 – 1,428,498 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -31.20 |
| Energy contribution | -32.33 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
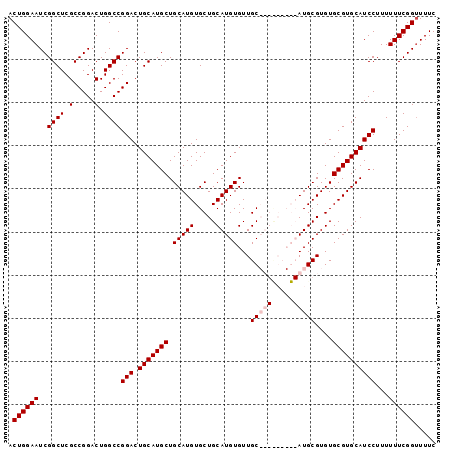
>X_DroMel_CAF1 1428399 99 + 22224390 ACUGGAAUCGGCUCGCCGGACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGCAUGCGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUC (((((((..((((.(.....).))))(((.((((((((((((((..((((((.....))))))..)))))))...))))))))))...))))))).... ( -41.90) >DroSec_CAF1 33173 90 + 1 ACUGGAAUCGGCUCGCCGGACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC---------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUC (((((((..((((.(.....).))))(((.(((((((.(((((((.((((.....)))---------))))))))))))))))))...))))))).... ( -36.10) >DroSim_CAF1 24717 90 + 1 ACUGGAAUCGGCUCGCCGGACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC---------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUC (((((((..((((.(.....).))))(((.(((((((.(((((((.((((.....)))---------))))))))))))))))))...))))))).... ( -36.10) >DroEre_CAF1 28398 99 + 1 ACUGGAAUCGGCUCGCCGGACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGCAUGUGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGAUUUC .((((((..((....))(((...((((.(((((((((.((((((((.((....)).)))))))).))))))).)).)).)).)))...))))))..... ( -40.90) >consensus ACUGGAAUCGGCUCGCCGGACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC_________AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUC .((((((..((((.(.....).))))(((.(((((((.(((((.....)))))...(((((.....)))))....))))))))))...))))))..... (-31.20 = -32.33 + 1.12)

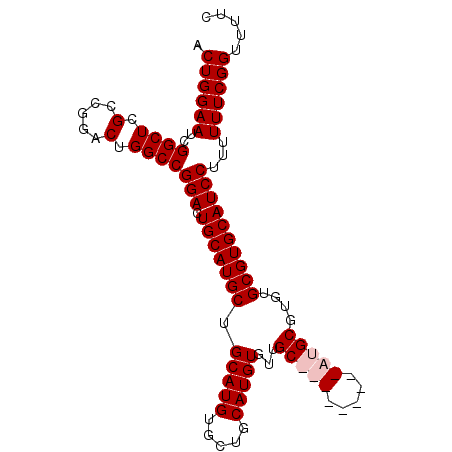
| Location | 1,428,399 – 1,428,498 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -23.90 |
| Energy contribution | -25.03 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1428399 99 - 22224390 GAAAACCGAAAAAAGGAUGCACGCACACGCAUGCAACGCAUGCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGUCCGGCGAGCCGAUUCCAGU ..............(((.((.(((....((((((...(((((.....))))).((....)).))))))....(((.....)))))).))....)))... ( -31.50) >DroSec_CAF1 33173 90 - 1 GAAAACCGAAAAAAGGAUGCACGCACACGCAU---------GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGUCCGGCGAGCCGAUUCCAGU ..............(((((((.((....((((---------(.....))))).((....)).)).))))...((((..(.....)..))))..)))... ( -25.70) >DroSim_CAF1 24717 90 - 1 GAAAACCGAAAAAAGGAUGCACGCACACGCAU---------GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGUCCGGCGAGCCGAUUCCAGU ..............(((((((.((....((((---------(.....))))).((....)).)).))))...((((..(.....)..))))..)))... ( -25.70) >DroEre_CAF1 28398 99 - 1 GAAAUCCGAAAAAAGGAUGCACGCACACGCAUGCAACACAUGCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGUCCGGCGAGCCGAUUCCAGU (.((((........(((((((.((....(((((.....)))))......((((.....)))))).)))).)))(((..(.....)..))))))).)... ( -28.60) >consensus GAAAACCGAAAAAAGGAUGCACGCACACGCAU_________GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGUCCGGCGAGCCGAUUCCAGU ..............(((((((.((....(((((.....)))))......((((.....)))))).))))...((((..(.....)..))))..)))... (-23.90 = -25.03 + 1.13)

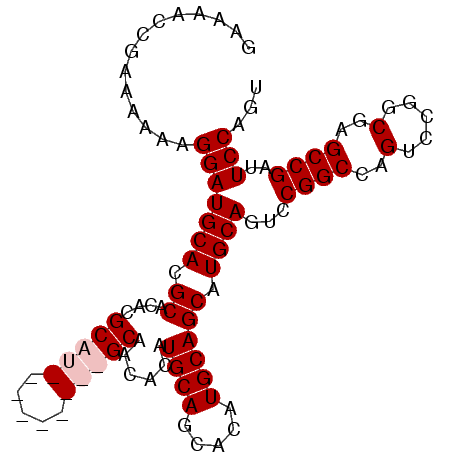
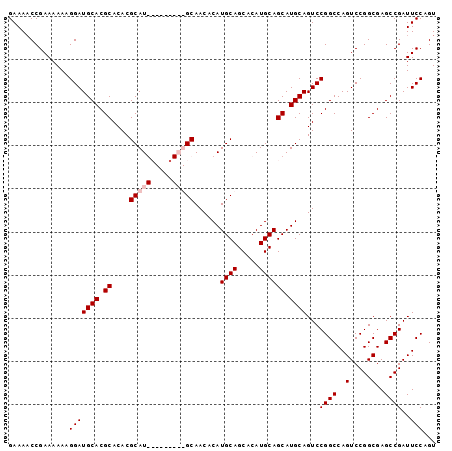
| Location | 1,428,418 – 1,428,537 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -43.01 |
| Consensus MFE | -37.78 |
| Energy contribution | -38.91 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1428418 119 + 22224390 ACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGCAUGCGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCACA ....((..(((.((((((.((....)).)))))))))((((((...))))))(((((((((..(((((((..((.....))..)).............))))))))))))))..))... ( -43.71) >DroSec_CAF1 33192 108 + 1 ACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC---------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA-- ....((......((((((.(((((((...))))))).))---------))))(((((((((..(((((((..((.....))..)).............))))))))))))))..)).-- ( -40.11) >DroSim_CAF1 24736 108 + 1 ACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC---------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA-- ....((......((((((.(((((((...))))))).))---------))))(((((((((..(((((((..((.....))..)).............))))))))))))))..)).-- ( -40.11) >DroEre_CAF1 28417 117 + 1 ACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGCAUGUGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGAUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA-- ....((......((((((.((((((((.((....)).)))))))).))))))((((((((((((((.......))))((((.(..(......)..).))))).)))))))))..)).-- ( -48.10) >consensus ACUGGCCGGACUGCAUGCUGCAUGUGCUGCAUGUGUUGC_________AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA__ ....((..(((.((((((.((....)).)))))))))(((((.....)))))(((((((((..(((((((..((.....))..)).............))))))))))))))..))... (-37.78 = -38.91 + 1.13)

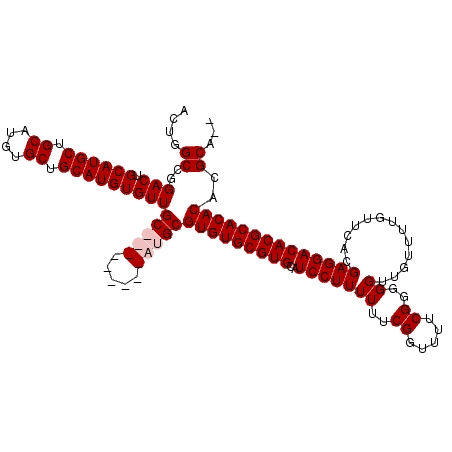
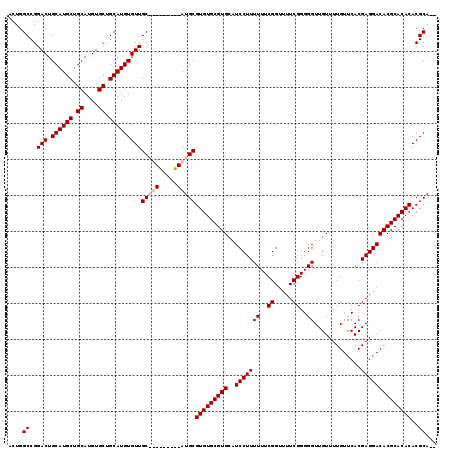
| Location | 1,428,418 – 1,428,537 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -36.35 |
| Energy contribution | -36.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1428418 119 - 22224390 UGUGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAUGCAACGCAUGCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGU ((.(((((((((.(((((((.(....)...........((.....)).....))))))).)))))))((((((...(((((.....))))).((....)).))))))......)))).. ( -42.60) >DroSec_CAF1 33192 108 - 1 --UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU---------GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGU --((((((((((.(((((((.(....)...........((.....)).....))))))).)))))))))).---------((.((.(((((.((....)).)))))..))...)).... ( -36.30) >DroSim_CAF1 24736 108 - 1 --UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU---------GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGU --((((((((((.(((((((.(....)...........((.....)).....))))))).)))))))))).---------((.((.(((((.((....)).)))))..))...)).... ( -36.30) >DroEre_CAF1 28417 117 - 1 --UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAUCCGAAAAAAGGAUGCACGCACACGCAUGCAACACAUGCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGU --.((..((((((((((....(....)...............((((.......))))))))))))))((((((......((((.....))))((....)).))))))......)).... ( -38.60) >consensus __UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU_________GCAACACAUGCAGCACAUGCAGCAUGCAGUCCGGCCAGU ..((((((((((.(((((((.(....)...........((.....)).....))))))).))))))))))..........((.((.(((((.((....)).)))))..))...)).... (-36.35 = -36.35 + 0.00)

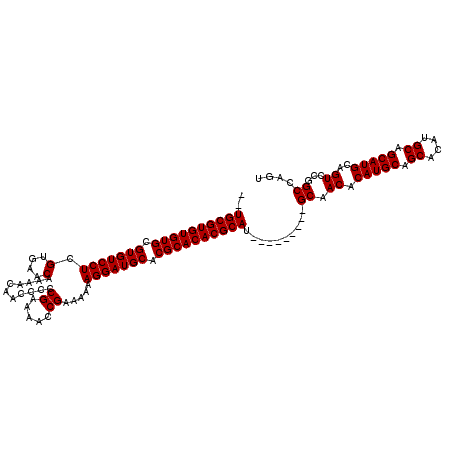
| Location | 1,428,458 – 1,428,557 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -25.63 |
| Energy contribution | -25.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1428458 99 + 22224390 UGCGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCACACAAACACAACGCACUGGCAA (((((((.....(((((((((...........(((((((((.(..(......)..).))))))).))....))))))))).....)))))))....... ( -32.76) >DroSec_CAF1 33231 83 + 1 --------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA--------CAACGCACUGGCAA --------.((((((((((((..(((((((..((.....))..)).............))))))))))))))..((.--------....))....))). ( -26.81) >DroSim_CAF1 24775 83 + 1 --------AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA--------CAACGCACUGGCAA --------.((((((((((((..(((((((..((.....))..)).............))))))))))))))..((.--------....))....))). ( -26.81) >DroEre_CAF1 28457 91 + 1 UGUGUUGCAUGCGUGUGCGUGCAUCCUUUUUUCGGAUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA--------CAACGCACUGGCAA (((((((..((.((((((((((((((.......))))((((.(..(......)..).))))).))))))))).))..--------)))))))....... ( -33.00) >consensus ________AUGCGUGUGCGUGCAUCCUUUUUUCGGUUUUCGGGGGUUGUUUUGUUCACGAGGACACGCACACACGCA________CAACGCACUGGCAA .........((((((((((((..(((((((..((.....))..)).............))))))))))))))..((.............))....))). (-25.63 = -25.63 + 0.00)

| Location | 1,428,458 – 1,428,557 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -27.45 |
| Energy contribution | -27.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1428458 99 - 22224390 UUGCCAGUGCGUUGUGUUUGUGUGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAUGCAACGCA .......((((((((......(((((((((((.(((((((.(....)...........((.....)).....))))))).))))))))))))))))))) ( -38.30) >DroSec_CAF1 33231 83 - 1 UUGCCAGUGCGUUG--------UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU-------- ..((....))...(--------((((((((((.(((((((.(....)...........((.....)).....))))))).)))))))))))-------- ( -28.40) >DroSim_CAF1 24775 83 - 1 UUGCCAGUGCGUUG--------UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU-------- ..((....))...(--------((((((((((.(((((((.(....)...........((.....)).....))))))).)))))))))))-------- ( -28.40) >DroEre_CAF1 28457 91 - 1 UUGCCAGUGCGUUG--------UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAUCCGAAAAAAGGAUGCACGCACACGCAUGCAACACA ..((....))((((--------(..((((((((((((....(....)...............((((.......))))))))))))))))..)))))... ( -32.90) >consensus UUGCCAGUGCGUUG________UGCGUGUGUGCGUGUCCUCGUGAACAAAACAACCCCCGAAAACCGAAAAAAGGAUGCACGCACACGCAU________ ..((....))............((((((((((.(((((((.(....)...........((.....)).....))))))).))))))))))......... (-27.45 = -27.45 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:06 2006