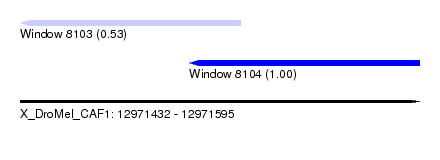
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,971,432 – 12,971,595 |
| Length | 163 |
| Max. P | 0.997817 |

| Location | 12,971,432 – 12,971,522 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12971432 90 - 22224390 AUUUUCAA--------AGUAACGAGCCAA---------------AACAGCAGAAGCCUGCAGAAGAUCUUGCAACGGUAAACUGGUUUGCAGGGCA---AAAACUCGACUGAAGUA ..(((((.--------.((..((((....---------------....((((....))))....(.(((((((.(((....)))...)))))))).---....))))))))))).. ( -20.40) >DroSim_CAF1 6434 91 - 1 AUUUUCAA--------AGUAACGAGCCAA--------------AAACAGCAGAAGCCUGCAGAAGAUCUUGCAACGGUAAACUGGUUUGCAGGGCA---AAAAGUCGACUGAAGUA ((((((..--------.((.....))...--------------.....((((....)))).))))))..(((..((((..(((..(((((...)))---)).)))..))))..))) ( -18.10) >DroEre_CAF1 5660 98 - 1 AUUUUCAAGCAUUUUCCGUAACAAGCCAA---------------AAAAGCAGAAGCCUGCAGAAGAUCUUGCAACGGUAAACUGGUUUGCAGUUCG---AAAAGUCGACUGGAAUA .....(((((..(((((((..((((....---------------....((((....)))).......))))..)))).)))...)))))((((.((---(....)))))))..... ( -22.26) >DroYak_CAF1 5967 116 - 1 AUCUCCAAGCAUUUUCAGUAACAAGCAAAAAAAACAAAACAAAAAAAAGCAGAAGCCUGCAGAAGAUCUUGCAACGGUAAACUGGUUUGCAGGGCACAAAAAAUUCGACUGAAAUA ............(((((((.............................((((....))))....(.(((((((.(((....)))...))))))))............))))))).. ( -22.20) >consensus AUUUUCAA________AGUAACAAGCCAA_______________AAAAGCAGAAGCCUGCAGAAGAUCUUGCAACGGUAAACUGGUUUGCAGGGCA___AAAAGUCGACUGAAAUA ((((((...........((.....))......................((((....)))).))))))((((((.(((....)))...))))))....................... (-14.96 = -15.27 + 0.31)

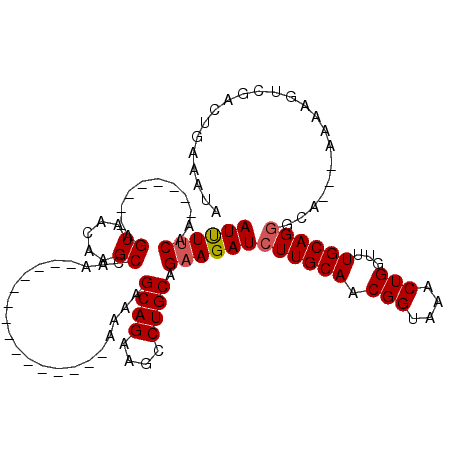
| Location | 12,971,501 – 12,971,595 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.10 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -11.42 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12971501 94 - 22224390 AA---AA----AGAAGAAGCAGAAAUGCUGCCAAAAACUAAAUGUUAAGUGGUAAAGUGAAACAAAAUUGCAUUUCGUGCAUUUUCAA--------AGUAACGAGCCAA----------- ..---..----.(((((.(((((((((((((((..(((.....)))...))))).(((........)))))))))).))).)))))..--------.............----------- ( -21.70) >DroSim_CAF1 6504 94 - 1 AA---AA----AGAAGAAGCAGAAACGCUGCCAAAAACUAAAUGUUAAGUGGUAAAGUGAAACAAAAUUGCAUUUCGUGCAUUUUCAA--------AGUAACGAGCCAA----------- ..---..----.(((((.(((((((.(((((((..(((.....)))...))))).(((........))))).)))).))).)))))..--------.............----------- ( -17.40) >DroEre_CAF1 5729 106 - 1 GA---AGACGCAGAAGAAGCAAUAAUGCUGCCAAAAAUUAAAUGUUAAGUGGUAAAGUGAAACAAAAUUGCAUUUUGUGCAUUUUCAAGCAUUUUCCGUAACAAGCCAA----------- ((---(((.((.(((((.(((..((((((((((..(((.....)))...))))).(((........))))))))...))).)))))..)).))))).............----------- ( -20.20) >DroYak_CAF1 6043 120 - 1 GAAGAAGAAGCAGAGGAAGCAGAAGUGCUGCCAAAAAUUAAAUGUUAAGUGGUAAAGUGAAACAAAAUUGCAUUUUGUGCAUCUCCAAGCAUUUUCAGUAACAAGCAAAAAAAACAAAAC ...(((((.((.(..((.(((((((((((((((..(((.....)))...))))).(((........)))))))))).))).))..)..)).)))))........................ ( -21.00) >DroAna_CAF1 8745 99 - 1 -------AGGCAGAA-AACCUCAAAGGCUGCCAAAAAUUAAAUGCUAAGUGGUAAAGCGAAACAGAAAUGCAUUUUGUUAC-UGCAAAAAAAUAAAAAUAAAAAACCA------------ -------.(((((..-..((.....)))))))................(..(((..((((((((....))..)))))))))-..).......................------------ ( -17.90) >consensus AA___AAA_GCAGAAGAAGCAGAAAUGCUGCCAAAAAUUAAAUGUUAAGUGGUAAAGUGAAACAAAAUUGCAUUUUGUGCAUUUUCAA__A_U___AGUAACAAGCCAA___________ ............(((((.(((((((((((((((..(((.....)))...)))))..(.....)......))))))).))).))))).................................. (-11.42 = -13.30 + 1.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:15 2006