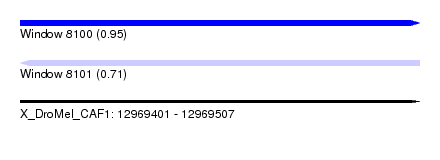
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,969,401 – 12,969,507 |
| Length | 106 |
| Max. P | 0.953238 |

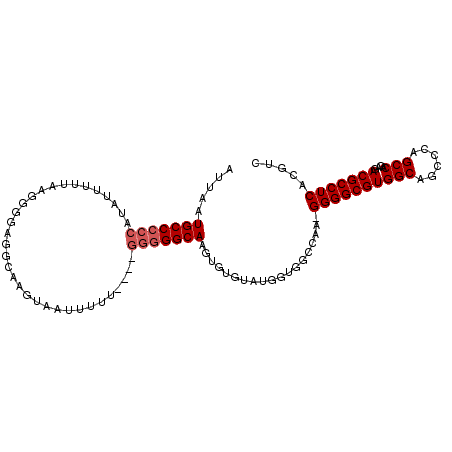
| Location | 12,969,401 – 12,969,507 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12969401 106 + 22224390 GACGUGAGGCGUCUCUGGCUGGGCUGCCACGCCCCCUUGGCCACCAUACACACCUGCCCCCCCCAAAAAAUUACUUGCCUCCCCUUAAAAAUAUGGGGGCAUUAAU ((((.....))))..(((((((((......))))....)))))...........(((((((.................................)))))))..... ( -29.61) >DroSim_CAF1 3700 93 + 1 GACGUGAGGCGUCUCUGGCUGGGCUGCCACGCCCC---------CAUACACACUUGCCCCC----AAAAAUUACUUGCCUCCCCUUAAAAAUAUGGGGGCAUUAAU ((((.....))))..(((..((((......)))))---------))........(((((((----(...........................))))))))..... ( -27.43) >DroEre_CAF1 3530 86 + 1 GACGUGAGGCGUCUGUGGCUGGGCUGCCACGCCCCUUUGGCCACCAUACAUUUGUGCC-----------------UGC---CCCCUAAAAAUAUGGGGGCAUUAAU ((((.....)))).((((((((((......))))....))))))..............-----------------(((---((((.........)))))))..... ( -35.10) >consensus GACGUGAGGCGUCUCUGGCUGGGCUGCCACGCCCC_UUGGCCACCAUACACACCUGCCCCC____AAAAAUUACUUGCCUCCCCUUAAAAAUAUGGGGGCAUUAAU ((((.....))))..((((......)))).((((((..........................................................))))))...... (-19.74 = -19.97 + 0.22)

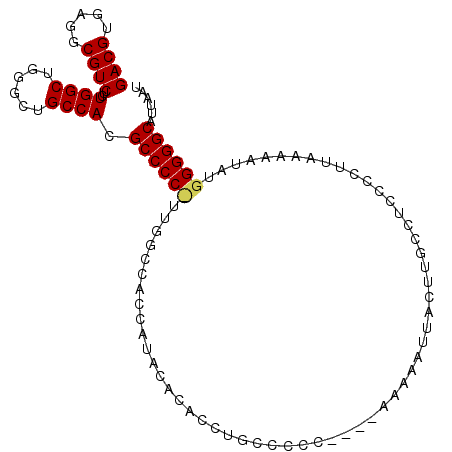

| Location | 12,969,401 – 12,969,507 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -23.24 |
| Energy contribution | -24.24 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12969401 106 - 22224390 AUUAAUGCCCCCAUAUUUUUAAGGGGAGGCAAGUAAUUUUUUGGGGGGGGCAGGUGUGUAUGGUGGCCAAGGGGGCGUGGCAGCCCAGCCAGAGACGCCUCACGUC .....(((((((...(((((....)))))((((......))))..)))))))(((((.(.((((..(....)((((......)))).)))).).)))))....... ( -36.00) >DroSim_CAF1 3700 93 - 1 AUUAAUGCCCCCAUAUUUUUAAGGGGAGGCAAGUAAUUUUU----GGGGGCAAGUGUGUAUG---------GGGGCGUGGCAGCCCAGCCAGAGACGCCUCACGUC .....((((((((.....(((...(....)...)))....)----))))))).......(((---------((((((((((......)))....))))))).))). ( -31.70) >DroEre_CAF1 3530 86 - 1 AUUAAUGCCCCCAUAUUUUUAGGGG---GCA-----------------GGCACAAAUGUAUGGUGGCCAAAGGGGCGUGGCAGCCCAGCCACAGACGCCUCACGUC .....(((((((.........))))---)))-----------------.......((((..(((((((.....)))(((((......)))))...))))..)))). ( -35.80) >consensus AUUAAUGCCCCCAUAUUUUUAAGGGGAGGCAAGUAAUUUUU____GGGGGCAAGUGUGUAUGGUGGCCAA_GGGGCGUGGCAGCCCAGCCAGAGACGCCUCACGUC .....(((((((.................................)))))))...................((((((((((......)))....)))))))..... (-23.24 = -24.24 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:13 2006