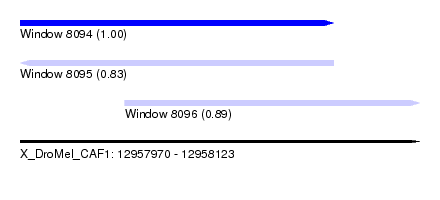
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,957,970 – 12,958,123 |
| Length | 153 |
| Max. P | 0.997907 |

| Location | 12,957,970 – 12,958,090 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.00 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12957970 120 + 22224390 AAUAUUUUACAUAUGCUUCGGAUGCACGUGUCCAACGCCACACGUGUCCUAAUUCUGUUAAUAAUACGUUGGCCAUUUUAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCGCAACCGG .............(((((.((((((((((((........))))))))........(((.....(((.(((((((((.....))))))))).)))...)))))))))))).(((....))) ( -32.00) >DroSec_CAF1 3376 120 + 1 AAUAUUUUACAUAUGCUUCGGAUGCACGUGUCCAACGACACACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAUUGGUCAACAUAUGCUACAAUUCAAGCAACCAGGCCCGG ..................(((..((((((((........)))))))).........(((........(((((((((.....)))))))))...((((........))))....)))))). ( -31.20) >DroSim_CAF1 2408 120 + 1 AAUAUUUUACAUAUGCUUCGGAUGCACGUGUCCAACGACACACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCAGACCCGG ..................(((..((((((((........))))))))......((((..........((((((((.......))))))))...((((........))))..)))).))). ( -30.90) >DroEre_CAF1 3525 119 + 1 GAUAUUUUACAUAUGCCUUGGAUACACGUAUCCAACGCCACACGUGCCCCAAUUCUGAUAAUAAUACGUUGGCUACUC-GAAUGGUCAGCAUAUGCUACAAUUCAAGCAACCAGAGCCGG ..............((.(((((((....))))))).))....((.((......((((..........((((((((...-...))))))))...((((........))))..)))))))). ( -27.80) >DroYak_CAF1 3504 112 + 1 AAUAAUUUACAUAUGCUGAGGAUACACGUGUCCCACGACACACGUGUCCCAAUUCUGUUAAUAAUACGUUGGCCAAGCUAAAUGGCCAACAUAUGCUACAAUUCAAGCAACC-------- .............((((((((((((..(((((....)))))..))))))......(((.....(((.((((((((.......)))))))).)))...)))..)).))))...-------- ( -33.10) >consensus AAUAUUUUACAUAUGCUUCGGAUGCACGUGUCCAACGACACACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCAGACCCGG .............(((((.((((((((((((........))))))))....................((((((((.......))))))))..........)))))))))........... (-25.36 = -25.56 + 0.20)



| Location | 12,957,970 – 12,958,090 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.00 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12957970 120 - 22224390 CCGGUUGCGGUUGCUUGAAUUGUAGCAUAUGUUGACCAUUUAAAAUGGCCAACGUAUUAUUAACAGAAUUAGGACACGUGUGGCGUUGGACACGUGCAUCCGAAGCAUAUGUAAAAUAUU ....(((((..(((((...((((...((((((((.((((.....)))).)))))))).....)))).....((((((((((........)))))))..))).)))))..)))))...... ( -33.20) >DroSec_CAF1 3376 120 - 1 CCGGGCCUGGUUGCUUGAAUUGUAGCAUAUGUUGACCAAUUUAGUUGGCCAACGUAUUAUUAGCAGAAUUAGGACACGUGUGUCGUUGGACACGUGCAUCCGAAGCAUAUGUAAAAUAUU .((((((((((((((........)))((((((((.(((((...))))).)))))))).........))))))).(((((((........)))))))..)))).................. ( -33.80) >DroSim_CAF1 2408 120 - 1 CCGGGUCUGGUUGCUUGAAUUGUAGCAUAUGUUGACCAUUUUAGUUGGCCAACGUAUUAUUAGCAGAAUUAGGACACGUGUGUCGUUGGACACGUGCAUCCGAAGCAUAUGUAAAAUAUU ((...(((((((((.......)))))((((((((.(((.......))).))))))))......))))....))..((((((((..(((((........))))).))))))))........ ( -33.90) >DroEre_CAF1 3525 119 - 1 CCGGCUCUGGUUGCUUGAAUUGUAGCAUAUGCUGACCAUUC-GAGUAGCCAACGUAUUAUUAUCAGAAUUGGGGCACGUGUGGCGUUGGAUACGUGUAUCCAAGGCAUAUGUAAAAUAUC ...(((((((((((((((((.(((((....))).)).))))-))))))))...(((....))).......)))))(((((((.(.(((((((....))))))).))))))))........ ( -43.40) >DroYak_CAF1 3504 112 - 1 --------GGUUGCUUGAAUUGUAGCAUAUGUUGGCCAUUUAGCUUGGCCAACGUAUUAUUAACAGAAUUGGGACACGUGUGUCGUGGGACACGUGUAUCCUCAGCAUAUGUAAAUUAUU --------.(((((.......)))))((((((((((((.......)))))))))))).....(((...((((((.(((((((((....))))))))).))).)))....)))........ ( -38.60) >consensus CCGGGUCUGGUUGCUUGAAUUGUAGCAUAUGUUGACCAUUUAAAUUGGCCAACGUAUUAUUAACAGAAUUAGGACACGUGUGUCGUUGGACACGUGCAUCCGAAGCAUAUGUAAAAUAUU ...........(((((...((((...((((((((.((((.....)))).)))))))).....)))).....((((((((((........)))))))..))).)))))............. (-25.34 = -25.70 + 0.36)


| Location | 12,958,010 – 12,958,123 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12958010 113 + 22224390 CACGUGUCCUAAUUCUGUUAAUAAUACGUUGGCCAUUUUAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCGCAACCGGGAAAAUCCUUCACCCACUAUGCCCAGCGCACUG ...((((.((.................(((((((((.....)))))))))...((((........))))...(((...(((...........)))....)))..)).)))).. ( -24.60) >DroSec_CAF1 3416 112 + 1 CACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAUUGGUCAACAUAUGCUACAAUUCAAGCAACCAGGCCCGGGAAAAUCCUUCACCCACUAUGACCA-UGCACUG ...((((.........(((........(((((((((.....)))))))))...((((........))))....)))..(((...........)))..........-.)))).. ( -23.90) >DroSim_CAF1 2448 112 + 1 CACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCAGACCCGGGAAAAUCCUUCACCCACUAUGACCA-UGCACUG ...((((.........((((((.....))))))........(((((((.....((((........)))).........(((...........)))....))))))-))))).. ( -23.70) >DroEre_CAF1 3565 111 + 1 CACGUGCCCCAAUUCUGAUAAUAAUACGUUGGCUACUC-GAAUGGUCAGCAUAUGCUACAAUUCAAGCAACCAGAGCCGGGAAAAUCCCACACCCACUUAGCCCA-GCCACUG ...(((......(((((..........((((((((...-...))))))))...((((........))))..)))))..(((.....))).....)))........-....... ( -23.40) >consensus CACGUGUCCUAAUUCUGCUAAUAAUACGUUGGCCAACUAAAAUGGUCAACAUAUGCUACAAUUCAAGCAACCAGACCCGGGAAAAUCCUUCACCCACUAUGACCA_UGCACUG ...((((......((((..........(((((((((.....)))))))))...((((........))))..))))...(((...........)))............)))).. (-21.67 = -21.80 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:08 2006