
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,953,535 – 12,953,748 |
| Length | 213 |
| Max. P | 0.928643 |

| Location | 12,953,535 – 12,953,640 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.60 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953535 105 + 22224390 UAAUUGGCAAAGUGUUUGUCAAUCUAUCCAGUCAACGGCAUUGCCACAAUGAUAUACCAUUGUCUG-GCCGUCAACGCUGCGUAUACGUAAUUUGUGUUGCACGUA ..((((((((.....))))))))....((((.....(((...)))((((((......)))))))))-).(((((((((((((....))))....)))))).))).. ( -28.50) >DroSec_CAF1 19786 94 + 1 UAAUUGGCAAAAAGUUUGUCAAUCUGUCCAGUC------------ACAAUGAUAUACCAUUGUCUGGGCCGUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGUA ..((((((((.....))))))))..((((((..------------((((((......))))))))))))(((((((((((((....))))....)))))).))).. ( -30.50) >DroEre_CAF1 11092 105 + 1 UAAUUGGCGAAAUAUUUGUCAAUCCGUCCAGUCGACCGCAUGGACACAAUGAUAGACCAUUGUCUG-GCCAUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGAC ..((((((((.....))))))))(((((((((.....)).)))))((((((......))))))..)-)....((((((((((....))))....))))))...... ( -25.70) >consensus UAAUUGGCAAAAUGUUUGUCAAUCUGUCCAGUC_AC_GCAU_G_CACAAUGAUAUACCAUUGUCUG_GCCGUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGUA ..((((((((.....))))))))......................((((((......))))))......(((((((((((((....))))....)))))).))).. (-19.72 = -19.83 + 0.11)



| Location | 12,953,575 – 12,953,680 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.14 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.53 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953575 105 + 22224390 UUGCCACAAUGAUAUACCAUUGUCUG-GCCGUCAACGCUGCGUAUACGUAAUUUGUGUUGCACGUAUACGCCCUGUUUGGCUGCCAGUUAAUGGCUGGGCGUGGCC ..(((((.........((((((.(((-((.(((((((..(((((((((((((....)))..))))))))))..)).))))).))))).))))))......))))). ( -42.36) >DroSec_CAF1 19819 100 + 1 -----ACAAUGAUAUACCAUUGUCUGGGCCGUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGU-UGGCUGCCAGUUAAUGGCUGGGCGUGGCC -----((((((......))))))...(((((((((((..((((((((((...((.....))))))))))))..)))-))))..(((((.....)))))....)))) ( -40.40) >DroEre_CAF1 11132 104 + 1 UGGACACAAUGAUAGACCAUUGUCUG-GCCAUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGACUACGCCCUGU-UGGCUGCCAGCUAAUGGCUGGGCGUGGCC .(((((..(((......))))))))(-(((..((((((((((....))))....)))))).......((((((.((-((((.....))))))....)))))))))) ( -36.10) >consensus U_G_CACAAUGAUAUACCAUUGUCUG_GCCGUCAACGCUGCGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGU_UGGCUGCCAGUUAAUGGCUGGGCGUGGCC .....((((((......))))))....(((((((((((((((....))))....)))))).))....((((((.........((((.....)))).))))))))). (-32.20 = -32.53 + 0.33)

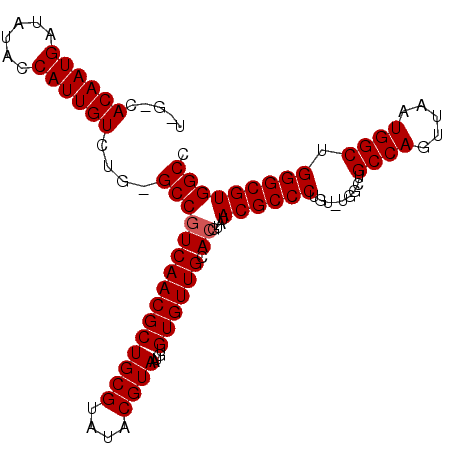

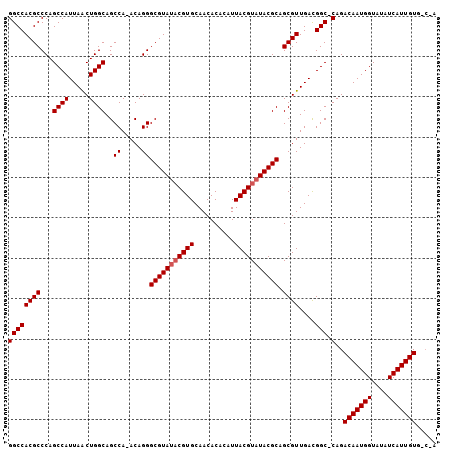
| Location | 12,953,575 – 12,953,680 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.14 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -31.57 |
| Energy contribution | -32.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953575 105 - 22224390 GGCCACGCCCAGCCAUUAACUGGCAGCCAAACAGGGCGUAUACGUGCAACACAAAUUACGUAUACGCAGCGUUGACGGC-CAGACAAUGGUAUAUCAUUGUGGCAA .((((((....((((((..(((((.(.(((.....(((((((((((..........)))))))))))....))).).))-)))..)))))).......)))))).. ( -41.30) >DroSec_CAF1 19819 100 - 1 GGCCACGCCCAGCCAUUAACUGGCAGCCA-ACAGGGCGUAUACGUGCAACACACAUUACGUAUACGCAGCGUUGACGGCCCAGACAAUGGUAUAUCAUUGU----- ((((.......((((.....)))).(.((-((...(((((((((((..........)))))))))))...)))).)))))...(((((((....)))))))----- ( -37.10) >DroEre_CAF1 11132 104 - 1 GGCCACGCCCAGCCAUUAGCUGGCAGCCA-ACAGGGCGUAGUCGUGCAACACACAUUACGUAUACGCAGCGUUGAUGGC-CAGACAAUGGUCUAUCAUUGUGUCCA (((...(((.(((.....)))))).))).-...(((((((((.(((.....))))))))))..((((((...(((((((-((.....))).)))))))))))))). ( -36.30) >consensus GGCCACGCCCAGCCAUUAACUGGCAGCCA_ACAGGGCGUAUACGUGCAACACACAUUACGUAUACGCAGCGUUGACGGC_CAGACAAUGGUAUAUCAUUGUG_C_A ((((((((...((((.....))))..((.....))(((((((((((..........))))))))))).))))....))).)..(((((((....)))))))..... (-31.57 = -32.23 + 0.67)


| Location | 12,953,614 – 12,953,720 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -36.38 |
| Energy contribution | -36.17 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953614 106 + 22224390 CGUAUACGUAAUUUGUGUUGCACGUAUACGCCCUGUUUGGCUGCCAGUUAAUGGCUGGGCGUGGCCAGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUGCAUGACUU .(((((((((((....)))..))))))))((..(((..((((((((.....)))).((((.(((((((...))))))).)))).))))....)))..))....... ( -37.50) >DroSec_CAF1 19854 105 + 1 CGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGU-UGGCUGCCAGUUAAUGGCUGGGCGUGGCCAGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUACAUGACUU .......((...((((((((((....(((((((...-.....((((.....)))).)))))))(((((...)))))(((((....))))).)))))))))).)).. ( -38.90) >DroEre_CAF1 11171 105 + 1 CGUAUACGUAAUGUGUGUUGCACGACUACGCCCUGU-UGGCUGCCAGCUAAUGGCUGGGCGUGGCCGAUUCUUGGCCAGCUCUUGGCCUGAUGCAAUGCAUGACUU .......((...((((((((((((.((((((((.((-((((.....))))))....)))))))).))..((..((((((...)))))).)))))))))))).)).. ( -42.90) >consensus CGUAUACGUAAUGUGUGUUGCACGUAUACGCCCUGU_UGGCUGCCAGUUAAUGGCUGGGCGUGGCCAGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUGCAUGACUU .......((...((((((((((....(((((((.........((((.....)))).)))))))(((((...)))))(((((....))))).)))))))))).)).. (-36.38 = -36.17 + -0.21)

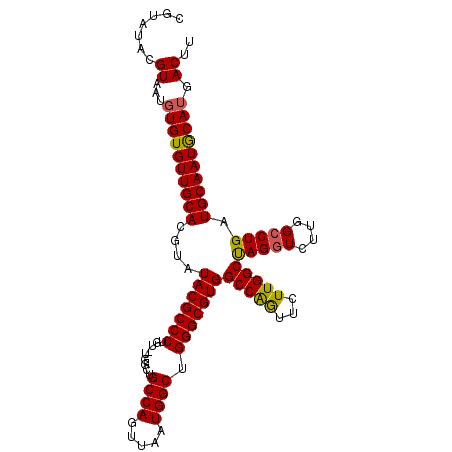
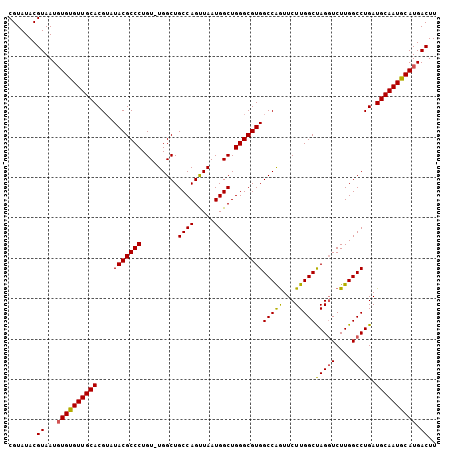
| Location | 12,953,614 – 12,953,720 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -28.70 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953614 106 - 22224390 AAGUCAUGCAUUGCAUCAGGCCAAGACCUAGCCAAGAACUGGCCACGCCCAGCCAUUAACUGGCAGCCAAACAGGGCGUAUACGUGCAACACAAAUUACGUAUACG .....((((...))))..(((((.(..((.....))..))))))..((((.((((.....)))).(.....).))))(((((((((..........))))))))). ( -30.40) >DroSec_CAF1 19854 105 - 1 AAGUCAUGUAUUGCAUCAGGCCAAGACCUAGCCAAGAACUGGCCACGCCCAGCCAUUAACUGGCAGCCA-ACAGGGCGUAUACGUGCAACACACAUUACGUAUACG ..((.((((.((((((..(((((.(..((.....))..))))))((((((.((((.....)))).(...-.).))))))....))))))...)))).))....... ( -32.20) >DroEre_CAF1 11171 105 - 1 AAGUCAUGCAUUGCAUCAGGCCAAGAGCUGGCCAAGAAUCGGCCACGCCCAGCCAUUAGCUGGCAGCCA-ACAGGGCGUAGUCGUGCAACACACAUUACGUAUACG ..((.(((..(((((...(((((.....)))))......((((.((((((.((((.....)))).(...-.).)))))).)))))))))....))).))....... ( -35.70) >consensus AAGUCAUGCAUUGCAUCAGGCCAAGACCUAGCCAAGAACUGGCCACGCCCAGCCAUUAACUGGCAGCCA_ACAGGGCGUAUACGUGCAACACACAUUACGUAUACG ......((..((((((..(((((....((.....))...)))))((((((.((((.....)))).(.....).))))))....))))))..))............. (-28.70 = -29.03 + 0.33)


| Location | 12,953,640 – 12,953,748 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.76 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -29.75 |
| Energy contribution | -30.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953640 108 + 22224390 UACGCCCUGUUUGGCUGCCAGUUAAUGGCUGGGCGUGGCCAGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUGCAUGACUUAAAUUUGACUGGGCU---AAGU---------AUAAAGUAG (((....((((((((..((((((((.(((..(((.(((((((...))))))).).))..)))..((((...)))).........)))))))))))---))))---------)....))). ( -37.30) >DroSec_CAF1 19880 107 + 1 UACGCCCUGU-UGGCUGCCAGUUAAUGGCUGGGCGUGGCCAGUUCUUGGCUAGGUCUUGGCCUGAUGCAAUACAUGACUUAAAUUUAACUGGGCG---AAGU---------AUAAAGUGG ..(((((.((-(((((((((.....)))).((((.(((((((...))))))).)))).))))..(((.....)))...........))).)))))---....---------......... ( -36.30) >DroEre_CAF1 11197 107 + 1 UACGCCCUGU-UGGCUGCCAGCUAAUGGCUGGGCGUGGCCGAUUCUUGGCCAGCUCUUGGCCUGAUGCAAUGCAUGACUUCAAUUUGACUGGGCG---AAGU---------ACAAAGUAU ..(((((.((-(((((((((.....)))).((((.(((((((...)))))))))))..))))..((((...))))...........))).)))))---....---------......... ( -40.80) >DroYak_CAF1 11601 119 + 1 UACGCCCCGU-UGACUGCCAGUUAAUGGCUGGGCGUGGCCGAUUCUUGGCCAGCUCUUGCCCUGAUGCAAUACAUGACUUCCAUUCAACUGGGCGGCAAAGUAUUGCGGCUAUAAAGUAG ..(((((.((-(((.((..((((...(((.((((.(((((((...)))))))))))..)))...(((.....)))))))..)).))))).)))))((((....)))).(((....))).. ( -43.70) >consensus UACGCCCUGU_UGGCUGCCAGUUAAUGGCUGGGCGUGGCCAAUUCUUGGCCAGCUCUUGGCCUGAUGCAAUACAUGACUUAAAUUUAACUGGGCG___AAGU_________AUAAAGUAG ..(((((.((..((((((((.....)))).((((.(((((((...))))))).)))).))))..(((.....)))............)).)))))......................... (-29.75 = -30.25 + 0.50)



| Location | 12,953,640 – 12,953,748 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.76 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953640 108 - 22224390 CUACUUUAU---------ACUU---AGCCCAGUCAAAUUUAAGUCAUGCAUUGCAUCAGGCCAAGACCUAGCCAAGAACUGGCCACGCCCAGCCAUUAACUGGCAGCCAAACAGGGCGUA .........---------....---.(.((((((...........((((...))))..(((.........)))..).))))).)((((((.((((.....)))).(.....).)))))). ( -26.30) >DroSec_CAF1 19880 107 - 1 CCACUUUAU---------ACUU---CGCCCAGUUAAAUUUAAGUCAUGUAUUGCAUCAGGCCAAGACCUAGCCAAGAACUGGCCACGCCCAGCCAUUAACUGGCAGCCA-ACAGGGCGUA .........---------....---.(.((((((....(((.(((.((..(((...)))..)).))).))).....)))))).)((((((.((((.....)))).(...-.).)))))). ( -26.90) >DroEre_CAF1 11197 107 - 1 AUACUUUGU---------ACUU---CGCCCAGUCAAAUUGAAGUCAUGCAUUGCAUCAGGCCAAGAGCUGGCCAAGAAUCGGCCACGCCCAGCCAUUAGCUGGCAGCCA-ACAGGGCGUA .........---------....---(((((........(((.(.((.....))).)))(((.....(((((((.......))))).))(((((.....)))))..))).-...))))).. ( -33.90) >DroYak_CAF1 11601 119 - 1 CUACUUUAUAGCCGCAAUACUUUGCCGCCCAGUUGAAUGGAAGUCAUGUAUUGCAUCAGGGCAAGAGCUGGCCAAGAAUCGGCCACGCCCAGCCAUUAACUGGCAGUCA-ACGGGGCGUA .............((((....))))(((((.(((((.(((..........((((......))))..(((((((.......))))).)))))((((.....))))..)))-)).))))).. ( -41.00) >consensus CUACUUUAU_________ACUU___CGCCCAGUCAAAUUGAAGUCAUGCAUUGCAUCAGGCCAAGACCUAGCCAAGAACCGGCCACGCCCAGCCAUUAACUGGCAGCCA_ACAGGGCGUA ..........................(((...((...........((((...))))..(((((.....)))))..))...))).((((((.((((.....)))).(.....).)))))). (-24.35 = -24.60 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:04 2006