
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,953,018 – 12,953,176 |
| Length | 158 |
| Max. P | 0.999582 |

| Location | 12,953,018 – 12,953,136 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -24.91 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
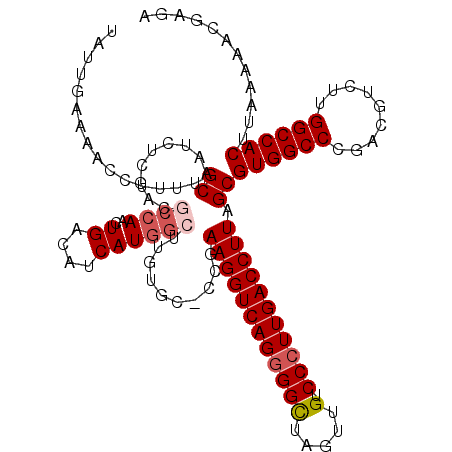
>X_DroMel_CAF1 12953018 118 + 22224390 UAGGGGGAUUGUGAAGGUAGAAGUCUUCGCUGGUUCAACC-AGCGUUUUUAAGUGGCCAAGACGUCGGGCCACGCUAAGGUCAAGGGACAACUAGCCCCUGACCUUCGGACACAACCCA ....(((.(((((..(((.........((((((.....))-)))).......((((((.........)))))))))(((((((.(((........))).)))))))....)))))))). ( -47.50) >DroSec_CAF1 19379 104 + 1 ---------------GGAAGAAGCCUUCGCUAGUUCAACGUCUCGUUUUUAAGUGGCCAAGACGUCGGGCCACGCUAAGGUCAAGGGACAACUAGCCGCUGACCUUCGGGCAAAAGCCA ---------------.((((.(((....(((((((....(((((..(((((.((((((.........))))))..)))))....)))))))))))).)))...)))).(((....))). ( -36.70) >DroEre_CAF1 10556 110 + 1 CAGGGGGCCCU---------CCACCUUCGGUGGGUCAACGUCUCGUUUUUAAGUGGCCAAGACGCGGGGCCACGCUAAGGUCAAGGGACAACCGGCCCCUGACCUUCGUGGACAAGCCA ..((.(((((.---------((......)).)))))...((((.........((((((.........))))))((.(((((((.(((........))).))))))).))))))...)). ( -45.30) >consensus _AGGGGG________GG_AGAAGCCUUCGCUGGUUCAACGUCUCGUUUUUAAGUGGCCAAGACGUCGGGCCACGCUAAGGUCAAGGGACAACUAGCCCCUGACCUUCGGGCACAAGCCA ......................((((..........................((((((.........))))))...(((((((.(((........))).))))))).))))........ (-24.91 = -25.47 + 0.56)


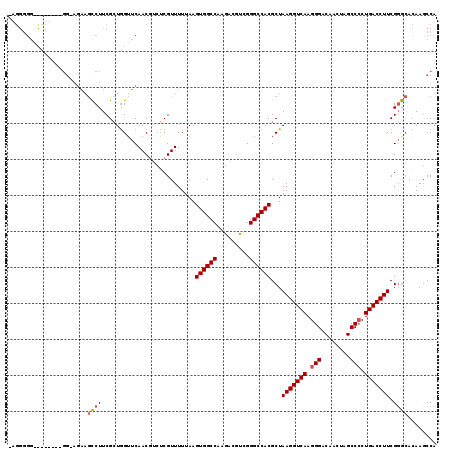
| Location | 12,953,018 – 12,953,136 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -29.66 |
| Energy contribution | -31.33 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953018 118 - 22224390 UGGGUUGUGUCCGAAGGUCAGGGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGCU-GGUUGAACCAGCGAAGACUUCUACCUUCACAAUCCCCCUA .((((((((....((((((((((((.....).)))))))))))...((((((.........)))))).......((((-((.....))))))..............))))))))..... ( -50.30) >DroSec_CAF1 19379 104 - 1 UGGCUUUUGCCCGAAGGUCAGCGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGAGACGUUGAACUAGCGAAGGCUUCUUCC--------------- .((((..(((...((((((((.(((.....).)).)))))))).))).)))).(((((((((............)))))))))(((..(((....)))..))).--------------- ( -39.20) >DroEre_CAF1 10556 110 - 1 UGGCUUGUCCACGAAGGUCAGGGGCCGGUUGUCCCUUGACCUUAGCGUGGCCCCGCGUCUUGGCCACUUAAAAACGAGACGUUGACCCACCGAAGGUGG---------AGGGCCCCCUG .(((((.(((((.(((((((((((.(....).))))))))))).(.((((..(.((((((((............)))))))).)..)))))....))))---------))))))..... ( -49.00) >consensus UGGCUUGUGCCCGAAGGUCAGGGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGAGACGUUGAACCAGCGAAGGCUUCU_CC________CCCCCU_ ..((((.......((((((((((((.....).)))))))))))..(((((..((((((((((............))))))))))..))).)).))))...................... (-29.66 = -31.33 + 1.67)


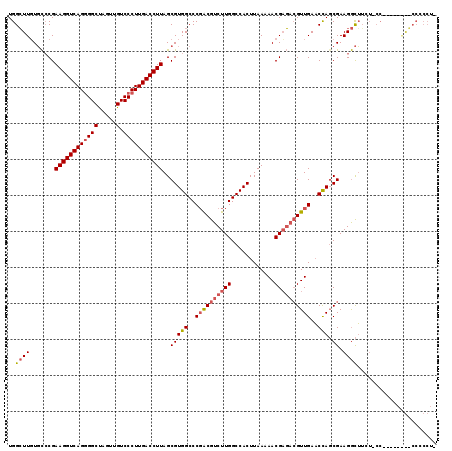
| Location | 12,953,058 – 12,953,176 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.68 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12953058 118 - 22224390 UAUUGAAAACCUUUUGCAAUCUGGACGCCAACUGACAUCAUGGGUUGUGU-CCGAAGGUCAGGGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGCU- ...............((....(((((((.((((.(.....).))))))))-)))((((((((((((.....).))))))))))).))((((((.........))))))...........- ( -43.50) >DroSec_CAF1 19404 119 - 1 UAUUGAAAACCUUUUGCAAUCUGGACGCCAACUGACAUCAUGGCUUUUGC-CCGAAGGUCAGCGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGAGA ......................(((((((..(((((.....(((....))-).....))))).))).....))))((((...((((.((((((.........))))))))))...)))). ( -32.90) >DroEre_CAF1 10587 119 - 1 UAUUGAAAACCUUUGGCAAUCUCGACGCCAACUGACAUCAUGGCUUGUCC-ACGAAGGUCAGGGGCCGGUUGUCCCUUGACCUUAGCGUGGCCCCGCGUCUUGGCCACUUAAAAACGAGA ...................(((((..((.....((((........)))).-...(((((((((((.(....).))))))))))).))((((((.........)))))).......))))) ( -39.10) >DroYak_CAF1 11092 93 - 1 UAUUGCGAACCUUUUGCAAUCUC------AGCUGACCUCAUGGCUCUUGGUCUGAAGGUCAGGGGUUAGUUGUCCCUUGACCUUAGC---------------------UUAAAAGCGAGC .(((((((.....)))))))(((------.(((((((...........))))..(((((((((((........)))))))))))...---------------------.....)))))). ( -33.00) >consensus UAUUGAAAACCUUUUGCAAUCUCGACGCCAACUGACAUCAUGGCUUGUGC_CCGAAGGUCAGGGGCUAGUUGUCCCUUGACCUUAGCGUGGCCCGACGUCUUGGCCACUUAAAAACGAGA ...............((.........((((..((....))))))..........((((((((((((.....).))))))))))).))((((((.........))))))............ (-24.24 = -25.68 + 1.44)

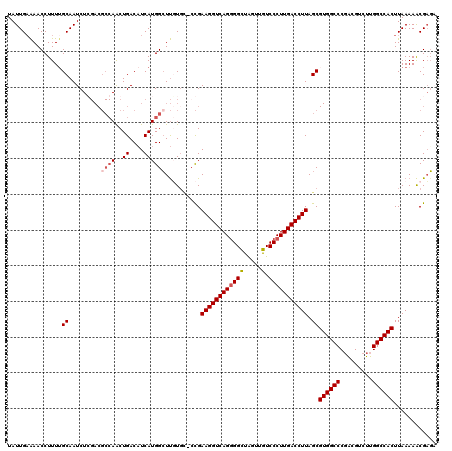
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:59 2006