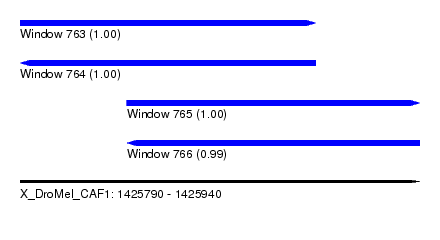
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,425,790 – 1,425,940 |
| Length | 150 |
| Max. P | 0.999773 |

| Location | 1,425,790 – 1,425,901 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.22 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -5.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1425790 111 + 22224390 UUUUUAACAUUUUAAAAAUGGCUGCCUUUUUGCACCACACGCACACACACAUACAUAUAUAUGUACUGCCAGUUUUCCGAGAGUGUGUGUGUGUGUGUGUGCGCA-AUCAGA (((((((....)))))))((..(((......(((((((((((((((((((.(((((....)))))((............)).))))))))))))))).)))))))-..)).. ( -38.90) >DroSec_CAF1 30519 105 + 1 UUUUUAACAUUUUAAAAAUGGCUGCCUUUUUGCACCACACGGACACACACA------CAUAUGUACUGCCAGUUUUCCGAGUGUUUGUGUGUGUCUGUGUGUGCA-UUCAGA .......((((.....)))).(((......(((((.(((((((((((((((------.((((..((.....)).......)))).))))))))))))))))))))-..))). ( -35.40) >DroAna_CAF1 40293 97 + 1 UUUUUAACAUUUUAAAAAUGGCUGCCUUUUUGCAUCAAACACACAUACACU------CAUAC--ACU-------CUCUUCGUAUGUGUGUGUGUGUGUGUAUGCAAUUCCGA .......((((.....)))).........((((((...((((((((((((.------(((((--...-------......))))).))))))))))))..))))))...... ( -31.80) >consensus UUUUUAACAUUUUAAAAAUGGCUGCCUUUUUGCACCACACGCACACACACA______CAUAUGUACUGCCAGUUUUCCGAGUGUGUGUGUGUGUGUGUGUGUGCA_UUCAGA .......((((.....))))..........(((((.((((((((((((((....................................)))))))))))))))))))....... (-26.88 = -26.22 + -0.66)

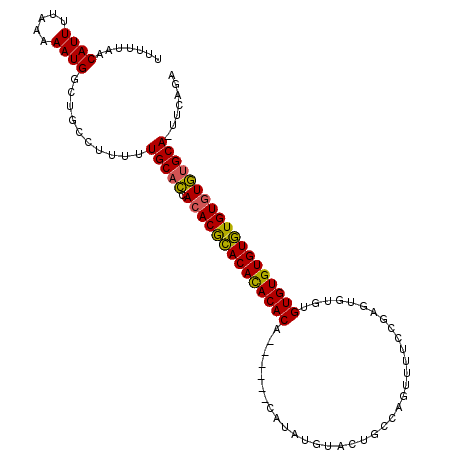
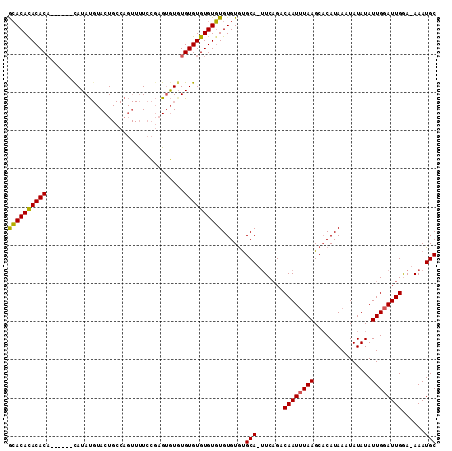
| Location | 1,425,790 – 1,425,901 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -23.06 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.05 |
| SVM RNA-class probability | 0.999773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1425790 111 - 22224390 UCUGAU-UGCGCACACACACACACACACACUCUCGGAAAACUGGCAGUACAUAUAUAUGUAUGUGUGUGUGCGUGUGGUGCAAAAAGGCAGCCAUUUUUAAAAUGUUAAAAA (((..(-(((((.(((((.((((((((((((.((((....)))).))(((((....))))))))))))))).)))))))))))..)))......(((((((....))))))) ( -39.90) >DroSec_CAF1 30519 105 - 1 UCUGAA-UGCACACACAGACACACACAAACACUCGGAAAACUGGCAGUACAUAUG------UGUGUGUGUCCGUGUGGUGCAAAAAGGCAGCCAUUUUUAAAAUGUUAAAAA (((...-(((((((((.(((((((((.......(((....)))(((.......))------)))))))))).)))).)))))...)))......(((((((....))))))) ( -31.20) >DroAna_CAF1 40293 97 - 1 UCGGAAUUGCAUACACACACACACACACAUACGAAGAG-------AGU--GUAUG------AGUGUAUGUGUGUUUGAUGCAAAAAGGCAGCCAUUUUUAAAAUGUUAAAAA ......((((((..(((((((.((((.((((((.....-------..)--)))))------.)))).)))))))...))))))...........(((((((....))))))) ( -27.70) >consensus UCUGAA_UGCACACACACACACACACACACACUCGGAAAACUGGCAGUACAUAUG______UGUGUGUGUGCGUGUGGUGCAAAAAGGCAGCCAUUUUUAAAAUGUUAAAAA .......(((((((((.(((((((((....................................))))))))).)))).)))))............(((((((....))))))) (-23.06 = -22.62 + -0.44)

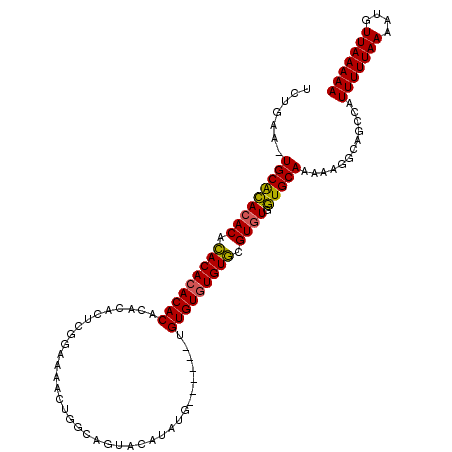
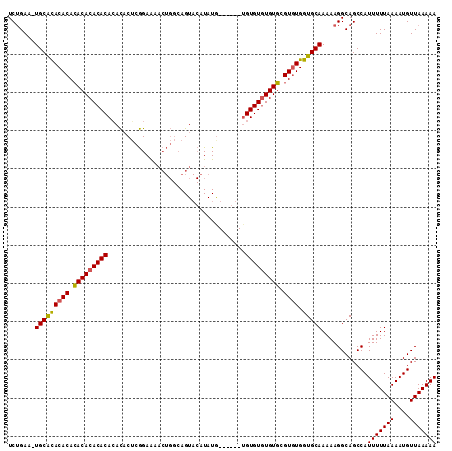
| Location | 1,425,830 – 1,425,940 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -29.01 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1425830 110 + 22224390 GCACACACACAUACAUAUAUAUGUACUGCCAGUUUUCCGAGAGUGUGUGUGUGUGUGUGUGCGCA-AUCAGACAAUUUAAGCACAUAAAUAUAUAUUGGAUUGGA-AAAUGC ((((((((((((((((((((......................))))))))))))))))))))(((-......((((((((...............))))))))..-...))) ( -35.51) >DroSec_CAF1 30559 104 + 1 GGACACACACA------CAUAUGUACUGCCAGUUUUCCGAGUGUUUGUGUGUGUCUGUGUGUGCA-UUCAGACAAUUUAAGCACAUAAAUAUAUAUUGGAUUGGA-AAAUGC ((....(((..------....)))....)).((((((((((((((((((((((((((.((....)-).))))).......))))))))))))........)))))-)))).. ( -25.91) >DroAna_CAF1 40333 97 + 1 ACACAUACACU------CAUAC--ACU-------CUCUUCGUAUGUGUGUGUGUGUGUGUAUGCAAUUCCGACAAUAUAAGCCCAUAAAUAUAUAUUGAAUUGAAAAAAUGC ((((((((((.------(((((--(..-------(.....)..)))))).))))))))))...((((((....((((((............))))))))))))......... ( -25.60) >consensus GCACACACACA______CAUAUGUACUGCCAGUUUUCCGAGUGUGUGUGUGUGUGUGUGUGUGCA_UUCAGACAAUUUAAGCACAUAAAUAUAUAUUGGAUUGGA_AAAUGC ((((((((((....................................))))))))))......(((.......((((((((...............))))))))......))) (-17.85 = -17.30 + -0.55)

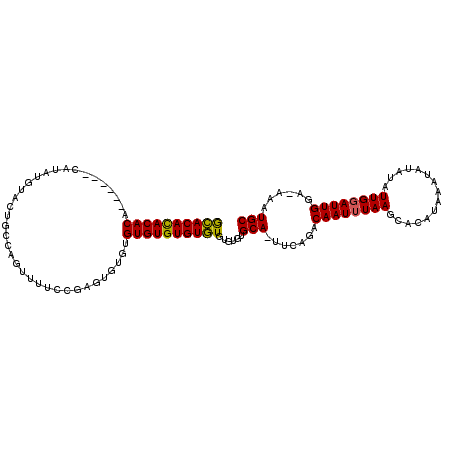
| Location | 1,425,830 – 1,425,940 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.74 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1425830 110 - 22224390 GCAUUU-UCCAAUCCAAUAUAUAUUUAUGUGCUUAAAUUGUCUGAU-UGCGCACACACACACACACACACUCUCGGAAAACUGGCAGUACAUAUAUAUGUAUGUGUGUGUGC (((.((-..((((...(((((....)))))......))))...)).-)))(((((((((((.......(((.((((....)))).)))((((....)))).))))))))))) ( -28.20) >DroSec_CAF1 30559 104 - 1 GCAUUU-UCCAAUCCAAUAUAUAUUUAUGUGCUUAAAUUGUCUGAA-UGCACACACAGACACACACAAACACUCGGAAAACUGGCAGUACAUAUG------UGUGUGUGUCC ((((((-..((((...(((((....)))))......))))...)))-))).......(((((((((.......(((....)))(((.......))------)))))))))). ( -24.80) >DroAna_CAF1 40333 97 - 1 GCAUUUUUUCAAUUCAAUAUAUAUUUAUGGGCUUAUAUUGUCGGAAUUGCAUACACACACACACACACAUACGAAGAG-------AGU--GUAUG------AGUGUAUGUGU (((....(((....(((((((............)))))))...))).)))......(((((.((((.((((((.....-------..)--)))))------.)))).))))) ( -23.80) >consensus GCAUUU_UCCAAUCCAAUAUAUAUUUAUGUGCUUAAAUUGUCUGAA_UGCACACACACACACACACACACACUCGGAAAACUGGCAGUACAUAUG______UGUGUGUGUGC (((......((((...(((((....)))))......)))).......))).......(((((((((....................................))))))))). (-14.52 = -14.74 + 0.22)

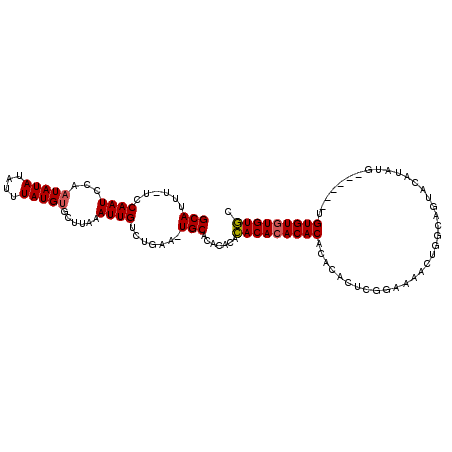
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:00 2006