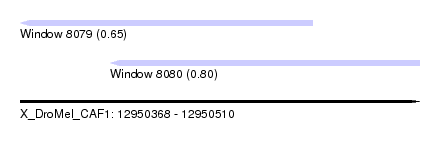
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,950,368 – 12,950,510 |
| Length | 142 |
| Max. P | 0.802076 |

| Location | 12,950,368 – 12,950,472 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -21.52 |
| Energy contribution | -24.02 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
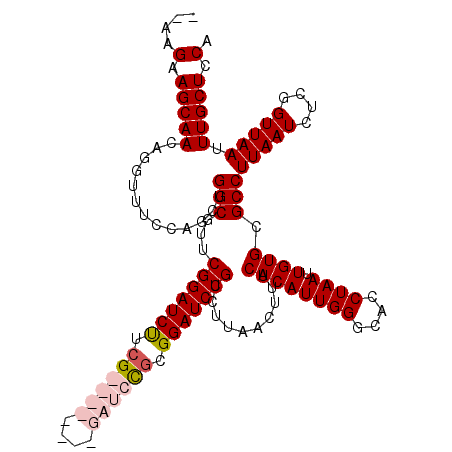
>X_DroMel_CAF1 12950368 104 - 22224390 --------GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCAGUUUUGAUCCGCGCUUGUGGCGCACAGAAA--------GC --------((((....))))........(((((..(((((....)))))((((((((.(((.(.(((((((...((....)).)))).))).).))).))))))))))))--------). ( -33.50) >DroSec_CAF1 8615 104 - 1 --------GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCAGUUUUGAUCCGCGCUUGCGGCGCACAGAAA--------GC --------((((....))))........(((((..(((((....)))))((((((((.(((.(.(((((((...((....)).)))).))).).))).))))))))))))--------). ( -33.50) >DroEre_CAF1 7956 112 - 1 GAUCUGCCGAUCUGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGGGCCUUAAUCUCGGUUAAUUUGCUCCAGUUUUGAUCCGCGCUUGUGGCGCACAGAAA--------GC (((((((......)))))))........(((((..(((((....)))))((((.(((.(((.(.(((((((...((....)).)))).))).).))).))).))))))))--------). ( -34.40) >DroYak_CAF1 8248 110 - 1 GAUCUUCGGAUCUUUGGAUCUGCCUUAACUUUCACAUUGGGCCCCUAAUUGUGGGCCUUAAUCUCGGUUAAUUUGCUCCAGUUUUGAUC----------ACACACAGAAAAGCCAGCUGC ((((....)))).((((.((((................(((((((.....).)))))).......((((((...((....)).))))))----------.....))))....)))).... ( -27.60) >consensus ________GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCAGUUUUGAUCCGCGCUUGUGGCGCACAGAAA________GC ........((((....)))).((((.............))))......(((((((((.(((.(.(((((((...((....)).)))).))).).))).)))))))))............. (-21.52 = -24.02 + 2.50)


| Location | 12,950,400 – 12,950,510 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -27.69 |
| Energy contribution | -29.50 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12950400 110 - 22224390 --AAGAAGCAACAGGUUUCCAGCUGGCGUUCGGAUCUUCG--------GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCA --..(.(((((..(((.((((((....)))((((((....--------)))))).)))...)))((((((........((((((......)))).))........)))))).))))).). ( -33.09) >DroSec_CAF1 8647 110 - 1 --AAGAAGCAACAGGUUUCCAGCUGGCGUUCGGAUCUUCG--------GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCA --..(.(((((..(((.((((((....)))((((((....--------)))))).)))...)))((((((........((((((......)))).))........)))))).))))).). ( -33.09) >DroEre_CAF1 7988 118 - 1 --AAGAAGCAACAGGUUUCCAGCUGGCGUUCGGAUCUUCGGAUCUGCCGAUCUGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGGGCCUUAAUCUCGGUUAAUUUGCUCCA --..(.(((((((((((..(((.((((....(((((....)))))))))..)))..))))))..((((((........((((.((.......)))))).......)))))).))))).). ( -37.66) >DroYak_CAF1 8278 120 - 1 AAAAGAAGCAACAGGUUUCCAACUGGCGUUCGGAUCCGCGGAUCUUCGGAUCUUUGGAUCUGCCUUAACUUUCACAUUGGGCCCCUAAUUGUGGGCCUUAAUCUCGGUUAAUUUGCUCCA ....(.((((((((........))).....((((((((.(((((....))))).))))))))..((((((........(((((((.....).)))))).......)))))).))))).). ( -42.16) >consensus __AAGAAGCAACAGGUUUCCAGCUGGCGUUCGGAUCUUCG________GAUCCGCGGAUCUGCCUUAACUUUCACAUUGGGCACCUAAUUGUGCGCCUUAAUCUCGGUUAAUUUGCUCCA ....(.(((((.............(((...(((((((.((((((....)))))).)))))))..........((((((((....)))).)))).)))(((((....))))).))))).). (-27.69 = -29.50 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:55 2006