
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,915,008 – 12,915,104 |
| Length | 96 |
| Max. P | 0.701986 |

| Location | 12,915,008 – 12,915,104 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -11.74 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
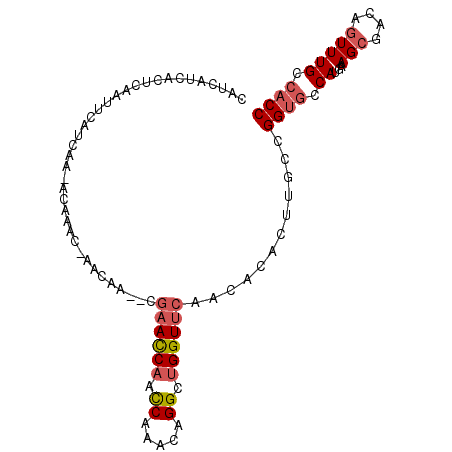
>X_DroMel_CAF1 12915008 96 - 22224390 CAUCAUCACUCAAUUCAUCAA-ACAAACAAACAA--CGAACCAACCAAACAGGCUGGUUCAACACUCUUGCCGGUGCCAUGAAGCGACAGUUUGCCACC .....................-.....(((((..--.((((((.((.....)).))))))..((((......)))).............)))))..... ( -17.30) >DroSec_CAF1 645 96 - 1 CAUCAUCACUCAAUUCAUCAA-ACAAACAAACAA--CGAACCAACCGAACAGGCUGGUUCAACACUCUUGCUGGUGCCAUGAAGCGACAGUUUGCCACC .....................-.....(((((..--.((((((.((.....)).)))))).......(((((..........)))))..)))))..... ( -19.20) >DroSim_CAF1 645 96 - 1 CAUCAUCACUCAAUUCAUCAA-ACAAACAAACAA--CGAACCAACCAAACAGGCUGGUUCAACACUCUUGCUGGUGCCAUGAAGCGACAGUUUGCCACC .....................-.....(((((..--.((((((.((.....)).)))))).......(((((..........)))))..)))))..... ( -18.50) >DroEre_CAF1 660 90 - 1 CAUCAUCACUCAAUCCAUCAA-ACAAA-----AA--CGAACC-ACCAAACAGGCUGGUUCAACACACUUGCCGGUGCCAUGAAGCGACAGUUUGCCACC ..................(((-((...-----..--.(((((-(((.....)).))))))....((((....)))).............)))))..... ( -16.20) >DroYak_CAF1 664 94 - 1 CAACAUCACUCAAUUCAUCAAAACAAA-----AAUGCGAACCAACCGAACAGGCUGGUUCAACACACUGGCCGGUGCCAUGAAGCGACAGUUUGCCACC .......................((((-----..(((((((((.((.....)).))))))....((.((((....))))))..)))....))))..... ( -20.30) >DroMoj_CAF1 806 78 - 1 -----------------CACA-UCAAUC-AUGAA--UCCAUCAAUCAAACAGGCUGGUUCAAUACACUCGCCGGCGCCAUGAAGCGACAAUUUGCGACC -----------------....-((..((-(((..--.........(.....)((((((...........))))))..))))).((((....)))))).. ( -13.00) >consensus CAUCAUCACUCAAUUCAUCAA_ACAAAC_AACAA__CGAACCAACCAAACAGGCUGGUUCAACACACUUGCCGGUGCCAUGAAGCGACAGUUUGCCACC .....................................((((((.((.....)).))))))............((((.((...(((....))))).)))) (-11.74 = -12.47 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:39 2006