
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,896,454 – 12,896,599 |
| Length | 145 |
| Max. P | 0.934930 |

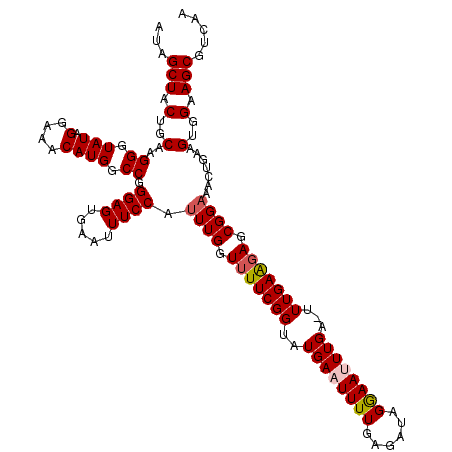
| Location | 12,896,454 – 12,896,570 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -30.11 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12896454 116 + 22224390 --GGCUACUGCAAGGGUAUAGGAAACAUGGCCGGGAGUGAAUUUCCAUUUGGUUUUCGGUAUGAAUUUUGAGAUAGAAAUUUGAAUUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAA --(((((((.(..((.(((.(....)))).))..)))))..(((((((((((((((((.(.(((((((.((........)).)))))))...).)))))))).))))))))).))).. ( -31.40) >DroSec_CAF1 97325 117 + 1 AUAGCUACUGCAAGGGUAUAGGAAACAUGGCCGGGAGUGAAUUUCCAUUUGGUUUUCGGUAUGAAUUUUUAGAUAGGAAUUUGA-UUUGAAGAGCGGAAACUAAAGUGGAAGCGUCAA ...(((.(..(..((.(((.(....)))).)).((((.....)))).(((((((((((.(........((((((....))))))-.......).))))))))))))..).)))..... ( -30.36) >DroSim_CAF1 81334 117 + 1 AUGGCUACUGCAAGGGUAUAGGAAACAUGGCCGGGAGUGAAUUUCCAUUUGGUUUUCGGUAUGAAUUUUGAGAUAGGAAUUUGA-UUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAA .((((((((.(..((.(((.(....)))).))..)))))..((((((((((.(((((((...(((((((......)))))))..-.))))))).)(....)..))))))))).)))). ( -31.40) >DroEre_CAF1 94422 117 + 1 AUAGCUACUGCAAGGGUAUGGAAAACAUGGCCGGGAGUGUAUUUCCAUUUGGUUUUCGGUUUGAUUUUUGAGAUAGAAAUUUGA-UUUGAGCAGCGGAAGCAGAAGUGGAAGCGUCAA ...((((((.(..((.((((.....)))).))..)))))...((((((((.((((..((((.((((((((...)))))))).))-)).)))).((....))..))))))))))..... ( -30.50) >DroYak_CAF1 105997 117 + 1 AUAGCUACUGCAAGGGUAUAGGAAACAUGGCCGGGAGUGUAUUUCCAUUUGGUUUUCGGUUUGAUUUUUGGGAUAGGAAUUUGA-UUUGAAGAGCGGCAAAAGAAGUGGAAGCGUCAA ...(((.(..(.........(....)...(((.((((.....))))....(.(((((((.(..(.((((......)))).)..)-.))))))).)))).......)..).)))..... ( -26.90) >consensus AUAGCUACUGCAAGGGUAUAGGAAACAUGGCCGGGAGUGAAUUUCCAUUUGGUUUUCGGUAUGAAUUUUGAGAUAGGAAUUUGA_UUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAA ...(((.(..(..((.(((.(....)))).)).((((.....)))).((((.(((((((..((((((((......))))))))...))))))).)))).......)..).)))..... (-24.56 = -24.96 + 0.40)

| Location | 12,896,492 – 12,896,599 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -14.65 |
| Energy contribution | -16.32 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
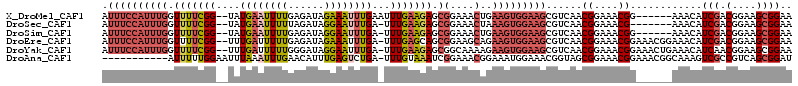
>X_DroMel_CAF1 12896492 107 + 22224390 AUUUCCAUUUGGUUUUCGG--UAUGAAUUUUGAGAUAGAAAUUUGAAUUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAACGGAAACGG------AAACAUCGACGGAAGCGGAA .(((((((((((((((((.--(.(((((((.((........)).)))))))...).)))))))).)))))))))((((...(....)(.------...)...))))......... ( -28.80) >DroSec_CAF1 97365 105 + 1 AUUUCCAUUUGGUUUUCGG--UAUGAAUUUUUAGAUAGGAAUUUGA-UUUGAAGAGCGGAAACUAAAGUGGAAGCGUCAACGGAAACG-------AAACAUCGACGGAAGCGGAA .(((((((((((((((((.--(........((((((....))))))-.......).)))))))).)))))))))......((....))-------.....(((.(....)))).. ( -26.66) >DroSim_CAF1 81374 106 + 1 AUUUCCAUUUGGUUUUCGG--UAUGAAUUUUGAGAUAGGAAUUUGA-UUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAACGGAAACGG------AAACAUCGACGGAAGCGGAA .((((((((((.(((((((--...(((((((......)))))))..-.))))))).)(....)..)))))))))((((...(....)(.------...)...))))......... ( -27.80) >DroEre_CAF1 94462 112 + 1 AUUUCCAUUUGGUUUUCGG--UUUGAUUUUUGAGAUAGAAAUUUGA-UUUGAGCAGCGGAAGCAGAAGUGGAAGCGUCAACGGAAACGGAAACGGAAACAUCGACGGAAGCGGAA .(((((((((.((((..((--((.((((((((...)))))))).))-)).)))).((....))..)))))))))((((..((....)).....(....)...))))......... ( -30.40) >DroYak_CAF1 106037 112 + 1 AUUUCCAUUUGGUUUUCGG--UUUGAUUUUUGGGAUAGGAAUUUGA-UUUGAAGAGCGGCAAAAGAAGUGGAAGCGUCAACGGAAACGGAAACUGAAACAUCAACGGAAGCGGAA .(((((..(((((((((((--(((......................-.((((.(..(.((.......)).)...).))))((....)).))))))))).))))).)))))..... ( -22.70) >DroAna_CAF1 108753 103 + 1 -----------AUUUUUGGAAUUUAAAUUUGAACAUUUGAGUCUGA-UUUGUAAAUCGGAAACGGAAAUGGAAACGGUAGCGGAAACGGAAACGGCAAAGUCGCCGUCAGCGGAU -----------....(..((.(((((((......)))))))))..)-(((((...(((....)))........((((..((.....((....)).....))..))))..))))). ( -24.30) >consensus AUUUCCAUUUGGUUUUCGG__UAUGAAUUUUGAGAUAGGAAUUUGA_UUUGAAGAGCGGAAACUGAAGUGGAAGCGUCAACGGAAACGG______AAACAUCGACGGAAGCGGAA .((((((((((.(((((((....((((((((......))))))))...))))))).)(....)..)))))))))......((....))............(((.(....)))).. (-14.65 = -16.32 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:37 2006