
| Sequence ID | X_DroMel_CAF1 |
|---|---|
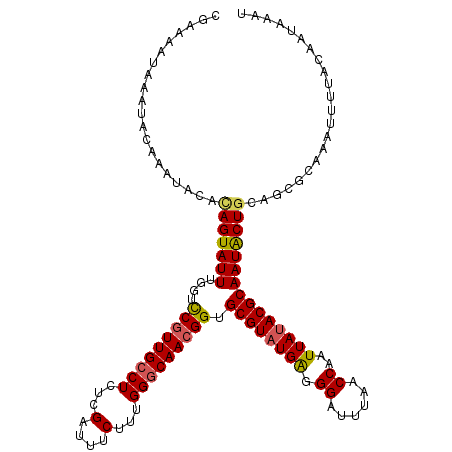
| Location | 12,864,171 – 12,864,291 |
| Length | 120 |
| Max. P | 0.971554 |

| Location | 12,864,171 – 12,864,291 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12864171 120 + 22224390 CGAAAAUAAGUACGAAUACAUAGUAUUUGGUCCGUUGCCUCUCGAUUUCUUUGGGCAACGGUGCGUAUGAAGGAUUUAACCAAUUAUACGCAAUACUGCAGCGCAACUUUUACAAUAAAU ((......(((((((((((...)))))))..(((((((((...(....)...)))))))))(((((((((.((......))..))))))))).))))....))................. ( -31.30) >DroSec_CAF1 62293 120 + 1 CGGAAAUAAAUACAAAUACACAGUAUUUGGUCCGUUGCCUCUCGAUUUCUUUGGGCAACGGUGCGUAUGAGGGAUUUAACCAAUUAUACGCAAUACUGCAGCGCAAAUUGUACAAUAAGU ............(((((((...)))))))..(((((((((...(....)...)))))))))(((((((((((.......))..)))))))))....(((((......)))))........ ( -31.60) >DroSim_CAF1 65578 120 + 1 CGGAAAUAAAUACAAAUACACAGUAUUUGGUCCGUUGCCUCUCGAUUUCUUUGGGCAACGGUGCGUAUGAGGGAUUUAACCAAUUAUACGCAAUACUGCAGCGCAAAUUGUACAAUAAGU ............(((((((...)))))))..(((((((((...(....)...)))))))))(((((((((((.......))..)))))))))....(((((......)))))........ ( -31.60) >DroEre_CAF1 66056 120 + 1 CGAAAAUAAAUACAAAUACACAGUAUUUGGUUCAUUGCCUCUCGGUUUCUUUGGCCAACGGUGCGUAUGAGGGAUUUAACCAAUUACACGCAAUGCUGCCACGCAAAUUUUACAACAAAC ..(((((...............(((.((((((...(.((((..((((.....)))).(((...)))..)))).)...)))))).)))......(((......))).)))))......... ( -21.60) >DroYak_CAF1 73770 120 + 1 CGAAAAGAAAUACAAAAACAGAGUAUUUGCUCCGUUGCCUCUCGAUUUCUUUGGGCAACGGUGCGUAUGGGGGAUUUAACCAAUUAUACGCAAUACUGCAGCGCAAAUUUUACAAUAAAU ......................((((((((.(((((((((...(....)...)))))))))((((((((.((.......))...))))))))..........)))))...)))....... ( -31.10) >consensus CGAAAAUAAAUACAAAUACACAGUAUUUGGUCCGUUGCCUCUCGAUUUCUUUGGGCAACGGUGCGUAUGAGGGAUUUAACCAAUUAUACGCAAUACUGCAGCGCAAAUUUUACAAUAAAU ....................(((((((....(((((((((...(....)...))))))))).((((((((.((......))..)))))))))))))))...................... (-26.10 = -26.30 + 0.20)

| Location | 12,864,171 – 12,864,291 |
|---|---|
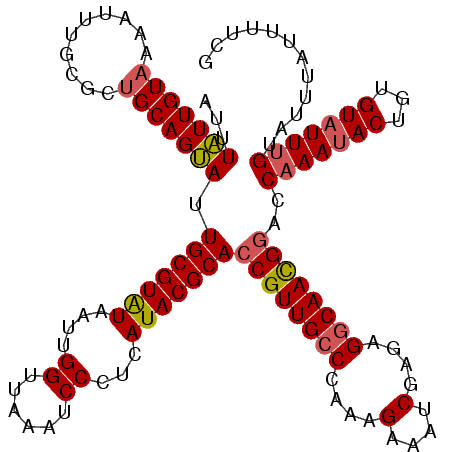
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -25.84 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12864171 120 - 22224390 AUUUAUUGUAAAAGUUGCGCUGCAGUAUUGCGUAUAAUUGGUUAAAUCCUUCAUACGCACCGUUGCCCAAAGAAAUCGAGAGGCAACGGACCAAAUACUAUGUAUUCGUACUUAUUUUCG ...........((((.(((.(((((((((((((((....((......))...)))))).((((((((....(....)....))))))))....)))))..))))..)))))))....... ( -31.80) >DroSec_CAF1 62293 120 - 1 ACUUAUUGUACAAUUUGCGCUGCAGUAUUGCGUAUAAUUGGUUAAAUCCCUCAUACGCACCGUUGCCCAAAGAAAUCGAGAGGCAACGGACCAAAUACUGUGUAUUUGUAUUUAUUUCCG .......((((((..(((...((((((((((((((....((......))...)))))).((((((((....(....)....))))))))....))))))))))).))))))......... ( -35.50) >DroSim_CAF1 65578 120 - 1 ACUUAUUGUACAAUUUGCGCUGCAGUAUUGCGUAUAAUUGGUUAAAUCCCUCAUACGCACCGUUGCCCAAAGAAAUCGAGAGGCAACGGACCAAAUACUGUGUAUUUGUAUUUAUUUCCG .......((((((..(((...((((((((((((((....((......))...)))))).((((((((....(....)....))))))))....))))))))))).))))))......... ( -35.50) >DroEre_CAF1 66056 120 - 1 GUUUGUUGUAAAAUUUGCGUGGCAGCAUUGCGUGUAAUUGGUUAAAUCCCUCAUACGCACCGUUGGCCAAAGAAACCGAGAGGCAAUGAACCAAAUACUGUGUAUUUGUAUUUAUUUUCG (((..(((.....(((...(((((((..(((((((....((......))...)))))))..))).))))..))).((....)))))..)))(((((((...)))))))............ ( -30.00) >DroYak_CAF1 73770 120 - 1 AUUUAUUGUAAAAUUUGCGCUGCAGUAUUGCGUAUAAUUGGUUAAAUCCCCCAUACGCACCGUUGCCCAAAGAAAUCGAGAGGCAACGGAGCAAAUACUCUGUUUUUGUAUUUCUUUUCG ............((((((..........(((((((....((......))...)))))))((((((((....(....)....)))))))).))))))........................ ( -29.40) >consensus AUUUAUUGUAAAAUUUGCGCUGCAGUAUUGCGUAUAAUUGGUUAAAUCCCUCAUACGCACCGUUGCCCAAAGAAAUCGAGAGGCAACGGACCAAAUACUGUGUAUUUGUAUUUAUUUUCG ...(((((((..........))))))).(((((((....((......))...)))))))((((((((....(....)....))))))))..(((((((...)))))))............ (-25.84 = -26.20 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:28 2006