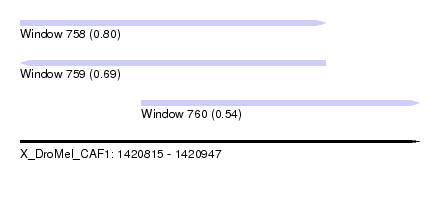
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,420,815 – 1,420,947 |
| Length | 132 |
| Max. P | 0.801561 |

| Location | 1,420,815 – 1,420,916 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -16.49 |
| Consensus MFE | -12.24 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1420815 101 + 22224390 UAUGACGAAUUUGUGCCCUAAAACUCUAAACGCUAGCUCAUUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCA .((((.(((.((((((...............))..(((((..(((......)))...)))))......)))).))).)))).....((....))....... ( -16.56) >DroSec_CAF1 27224 101 + 1 UAUGACGAAUUUGUGCCCUAAAACUCUAAACGAUAGCUCAUUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCA .((((.(((.((((....((......))...(((((((((..(((......)))...))))).)))).)))).))).)))).....((....))....... ( -18.00) >DroSim_CAF1 17865 101 + 1 UAUGACGAGUUUGUGCCCUAAAACUCUAAACGCUAGCUCAUUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCA ......((((((........)))))).....(((((((((..(((......)))...)))))........................((....))..)))). ( -21.50) >DroEre_CAF1 22502 82 + 1 UAUGACGAAUUUGUGCCCUAAAACUCU-------AGCUCAUUUGCACACACGCACACUCAGCAUAUCAACAACUUCAUCAUCAUU------------AGCA .((((.(((.((((.............-------.((......))......((.......))......)))).))).))))....------------.... ( -9.90) >consensus UAUGACGAAUUUGUGCCCUAAAACUCUAAACGCUAGCUCAUUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCA .((((.(((.(((((........)...........(((((..(((......)))...)))))......)))).))).)))).................... (-12.24 = -12.30 + 0.06)

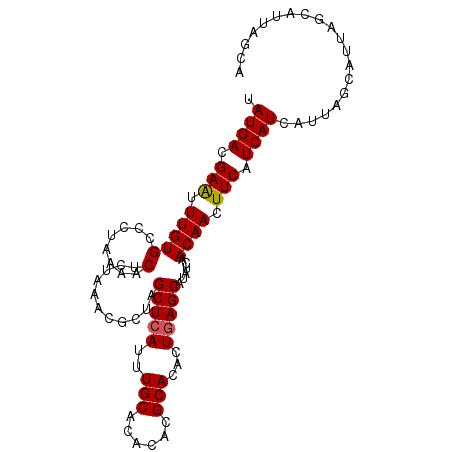
| Location | 1,420,815 – 1,420,916 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1420815 101 - 22224390 UGCUAAUGCUAAUGCUAAUGAUGAUGAAGUUGUUGAUAUGCUCAGUGUGCGUGUGUGCAAAUGAGCUAGCGUUUAGAGUUUUAGGGCACAAAUUCGUCAUA .((....))...........((((((((.((((......(((((...((((....))))..)))))..((.(((((....))))))))))).)))))))). ( -24.80) >DroSec_CAF1 27224 101 - 1 UGCUAAUGCUAAUGCUAAUGAUGAUGAAGUUGUUGAUAUGCUCAGUGUGCGUGUGUGCAAAUGAGCUAUCGUUUAGAGUUUUAGGGCACAAAUUCGUCAUA .((....))...........((((((((.((((.((((.(((((...((((....))))..))))))))).......(((....))))))).)))))))). ( -27.00) >DroSim_CAF1 17865 101 - 1 UGCUAAUGCUAAUGCUAAUGAUGAUGAAGUUGUUGAUAUGCUCAGUGUGCGUGUGUGCAAAUGAGCUAGCGUUUAGAGUUUUAGGGCACAAACUCGUCAUA ....((((((((((.(((..((......))..))).)))(((((...((((....))))..))))))))))))..((((((........))))))...... ( -24.50) >DroEre_CAF1 22502 82 - 1 UGCU------------AAUGAUGAUGAAGUUGUUGAUAUGCUGAGUGUGCGUGUGUGCAAAUGAGCU-------AGAGUUUUAGGGCACAAAUUCGUCAUA ....------------....((((((((.......((((((.......))))))((((....((((.-------...))))....))))...)))))))). ( -22.30) >consensus UGCUAAUGCUAAUGCUAAUGAUGAUGAAGUUGUUGAUAUGCUCAGUGUGCGUGUGUGCAAAUGAGCUAGCGUUUAGAGUUUUAGGGCACAAAUUCGUCAUA ....................((((((((.......((((((.......))))))((((....(((((.........)))))....))))...)))))))). (-21.60 = -21.85 + 0.25)

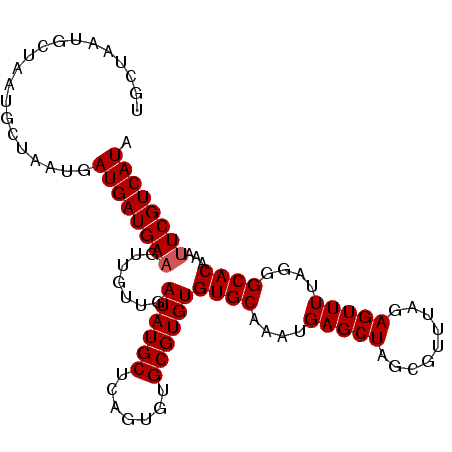
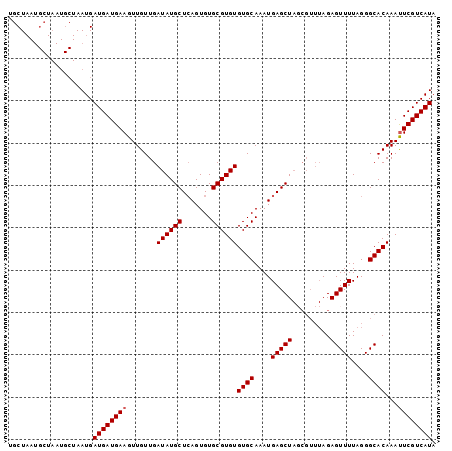
| Location | 1,420,855 – 1,420,947 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -16.57 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1420855 92 + 22224390 UUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCACGCAGCAGUUAUGCUCCUGUGUUAGCCCUAA ...........((((((.(((((((.(...................((....))....((.....)).).)))))))..))))..))..... ( -17.50) >DroSec_CAF1 27264 92 + 1 UUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCACGCAGCAGUUAUGCUCUUGUGUUAGCCCUAA ...........(((((..(((((((.(...................((....))....((.....)).).))))))).)))))......... ( -17.80) >DroSim_CAF1 17905 92 + 1 UUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCACGCAGCAGUUAUGCUCUUGUGUUAGCCUUAA ...........(((((..(((((((.(...................((....))....((.....)).).))))))).)))))......... ( -17.80) >DroEre_CAF1 22535 80 + 1 UUUGCACACACGCACACUCAGCAUAUCAACAACUUCAUCAUCAUU------------AGCACGAAACAGUUGUGCUCUUGUGUUAGCCCUAA ...((..((((((.......)).......................------------(((((((.....)))))))...))))..))..... ( -13.20) >consensus UUUGCACACACGCACACUGAGCAUAUCAACAACUUCAUCAUCAUUAGCAUUAGCAUUAGCACGCAGCAGUUAUGCUCUUGUGUUAGCCCUAA ...........(((((..(((((((.(...................((....))....((.....)).).))))))).)))))......... (-12.64 = -13.07 + 0.44)

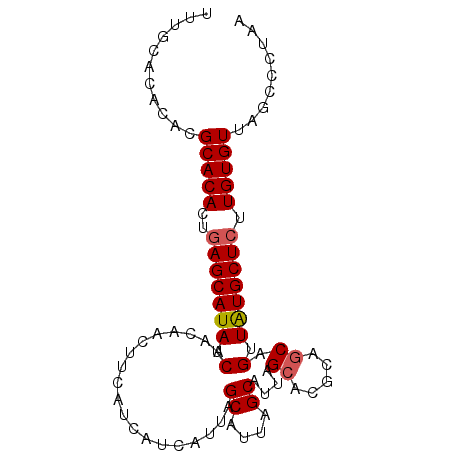
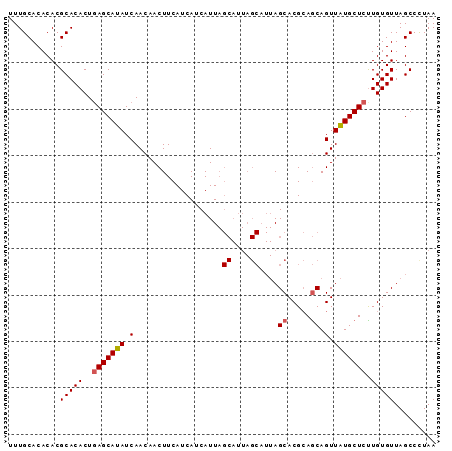
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:55 2006