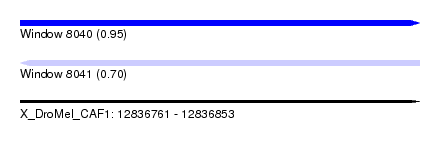
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,836,761 – 12,836,853 |
| Length | 92 |
| Max. P | 0.951855 |

| Location | 12,836,761 – 12,836,853 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.60 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.47 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12836761 92 + 22224390 AUUUUAUGUUUUUUGCUUUUUU-----U---UUUUUUUUGAA-GAGGCCUCAAA----GAGCGCUUUGAUUUUCUCACGCCGAUAAAAUCAAAGGCUCGAAAGAC .......(((((((((((((((-----.---........)))-)))))......----((((.((((((((((.((.....)).))))))))))))))))))))) ( -26.60) >DroPse_CAF1 56372 91 + 1 CGCGUC---GUUCUGUUGUCAC--------UCUUCCAUUGAACGUGUCUUCAAAGUGUGAGGGCUUUGAUUUUCUUACGACGAUAAAAUCAAAGU---AAAAGAC ..((((---((......(((((--------(((..(((((((......))))..)))..))))...))))......)))))).............---....... ( -18.90) >DroSim_CAF1 44760 91 + 1 AUUUUAUGUAUUUUGUUUUUUC---------UUUUUUUUGAA-GAGGCCUCAAA----GAGCGCUUUGAUUUUCUCACGCCGAUAAAAUCAAAGGCUCGACAGAC ......(((.....((((((((---------........)))-)))))......----((((.((((((((((.((.....)).)))))))))))))).)))... ( -25.00) >DroWil_CAF1 40024 90 + 1 AUUGAG--------GUUGUCACUUUGACGUUGCAUGUUUGAG-GGCGCUUCAAA----GUUAACUUUGAUUUUUUCACGCCGAUAAAAUCAAAGCC--CAAAGAC .(((((--------((.(((.(((.(((((...))))).)))-)))))))))).----.....((((((((((.((.....)).))))))))))..--....... ( -24.00) >DroYak_CAF1 43500 77 + 1 AUUUA----AU-------------------UUUUUUCUUGAA-GAGGCCUCAAA----GAGCGCUUUGAUUUUCACACGCCGAUAAAAUCAAAGGCUCGCAAGAC .....----..-------------------.....(((((..-((....))...----((((.((((((((((..(.....)..)))))))))))))).))))). ( -19.30) >DroPer_CAF1 53804 91 + 1 CGCGUC---GUUCUGUUGUCAC--------UCUUCCAUUGAACGUGUCUUCAAAGUGUGAGGGCUUUGAUUUUCUUACGACGAUAAAAUCAAAGU---AAAAGAC ..((((---((......(((((--------(((..(((((((......))))..)))..))))...))))......)))))).............---....... ( -18.90) >consensus AUUGUA___AUU_UGUUGUCAC________UUUUUUUUUGAA_GAGGCCUCAAA____GAGCGCUUUGAUUUUCUCACGCCGAUAAAAUCAAAGGC__GAAAGAC .....................................((((........))))..........((((((((((.((.....)).))))))))))........... ( -9.52 = -9.47 + -0.05)



| Location | 12,836,761 – 12,836,853 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.60 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -8.22 |
| Energy contribution | -8.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12836761 92 - 22224390 GUCUUUCGAGCCUUUGAUUUUAUCGGCGUGAGAAAAUCAAAGCGCUC----UUUGAGGCCUC-UUCAAAAAAAA---A-----AAAAAAGCAAAAAACAUAAAAU (((((..((((((((((((((.((.....)).)))))))))).))))----...)))))...-...........---.-----...................... ( -21.70) >DroPse_CAF1 56372 91 - 1 GUCUUUU---ACUUUGAUUUUAUCGUCGUAAGAAAAUCAAAGCCCUCACACUUUGAAGACACGUUCAAUGGAAGA--------GUGACAACAGAAC---GACGCG ((((((.---.(((((((((.....((....)))))))))))............)))))).(((((..((.....--------....))...))))---)..... ( -16.64) >DroSim_CAF1 44760 91 - 1 GUCUGUCGAGCCUUUGAUUUUAUCGGCGUGAGAAAAUCAAAGCGCUC----UUUGAGGCCUC-UUCAAAAAAAA---------GAAAAAACAAAAUACAUAAAAU .(((...((((((((((((((.((.....)).)))))))))).))))----(((((((...)-))))))....)---------)).................... ( -21.90) >DroWil_CAF1 40024 90 - 1 GUCUUUG--GGCUUUGAUUUUAUCGGCGUGAAAAAAUCAAAGUUAAC----UUUGAAGCGCC-CUCAAACAUGCAACGUCAAAGUGACAAC--------CUCAAU (((....--((((((((((((.((.....)).)))))))))))).((----(((((.(((..-........)))....))))))))))...--------...... ( -21.50) >DroYak_CAF1 43500 77 - 1 GUCUUGCGAGCCUUUGAUUUUAUCGGCGUGUGAAAAUCAAAGCGCUC----UUUGAGGCCUC-UUCAAGAAAAAA-------------------AU----UAAAU (((((..((((((((((((((...........)))))))))).))))----...)))))...-............-------------------..----..... ( -18.80) >DroPer_CAF1 53804 91 - 1 GUCUUUU---ACUUUGAUUUUAUCGUCGUAAGAAAAUCAAAGCCCUCACACUUUGAAGACACGUUCAAUGGAAGA--------GUGACAACAGAAC---GACGCG ((((((.---.(((((((((.....((....)))))))))))............)))))).(((((..((.....--------....))...))))---)..... ( -16.64) >consensus GUCUUUC__GCCUUUGAUUUUAUCGGCGUGAGAAAAUCAAAGCGCUC____UUUGAAGCCUC_UUCAAAAAAAAA________GUGACAACA_AAC___GAAAAU ...........((((((((((.((.....)).))))))))))..........(((((......)))))..................................... ( -8.22 = -8.72 + 0.50)

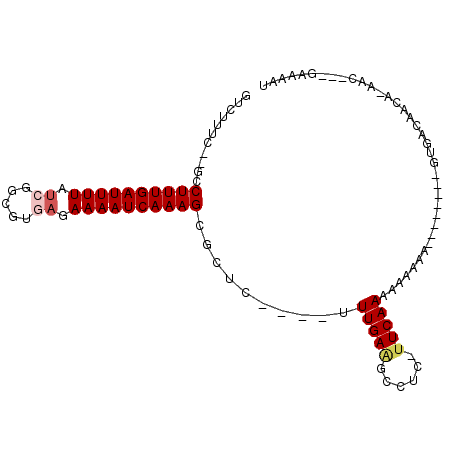

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:19 2006