
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,834,746 – 12,834,837 |
| Length | 91 |
| Max. P | 0.999732 |

| Location | 12,834,746 – 12,834,837 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -26.70 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
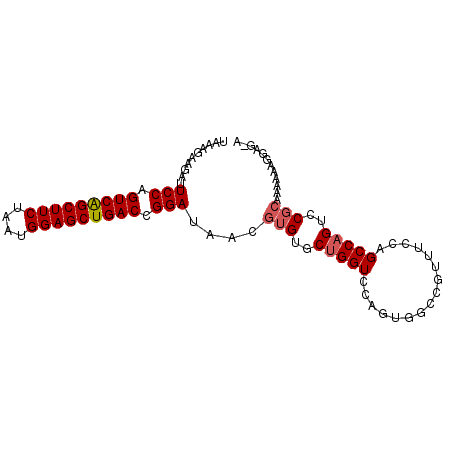
>X_DroMel_CAF1 12834746 91 + 22224390 U-CUCCUUUUUUGCGGACUGGCUGGAAACGGUCACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCUGACUGGAAUCUUCUUUA .-.........(((((.(((((((....))))))..)..)).))).......(((((((((((.(....).))))))))))).......... ( -31.60) >DroSec_CAF1 34314 91 + 1 U-CUCCUUUUUUGCGGACUGGCUGGAAACGGCCACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCCGACUGGAAUCUUCUUUA .-.........(((((.(((((((....))))))..)..)).))).......(((((((.(((.(....).))).))))))).......... ( -29.70) >DroSim_CAF1 42721 91 + 1 U-CUCCUUUUUUGCGGACUGGCUGGAAACGGCCACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCUGACUGGAAUCUUCUUUA .-.........(((((.(((((((....))))))..)..)).))).......(((((((((((.(....).))))))))))).......... ( -34.30) >DroEre_CAF1 39489 88 + 1 --CGCCUUUUUU--GGACAGGCUUGAAAUGGCUACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCUGACUGGAGUCUUCUUUA --.(((.((((.--((.....)).)))).)))....((((.(((....))).(((((((((((.(....).)))))))))))))))...... ( -27.60) >DroYak_CAF1 41501 92 + 1 UCCACUUUUUUUGCGGACUGGCUUGAAAUGGCUACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCUGACUGGAAUCUUCUUUA ...........(((((.((((((......))))...)).)).))).......(((((((((((.(....).))))))))))).......... ( -26.60) >consensus U_CUCCUUUUUUGCGGACUGGCUGGAAACGGCCACUGGACCAGCACACGUUAUCCGGUCAGCUCCAUUAGAAGCUGACUGGAAUCUUCUUUA ...........(((((.(((((((....))))))..)..)).))).......(((((((((((.(....).))))))))))).......... (-26.70 = -27.50 + 0.80)



| Location | 12,834,746 – 12,834,837 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -27.37 |
| Energy contribution | -27.81 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12834746 91 - 22224390 UAAAGAAGAUUCCAGUCAGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUGACCGUUUCCAGCCAGUCCGCAAAAAAGGAG-A .........((((.(((((((((....)))))))))(((((....((.(((((....(....)....)))))))))))).......))))-. ( -31.70) >DroSec_CAF1 34314 91 - 1 UAAAGAAGAUUCCAGUCGGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUGGCCGUUUCCAGCCAGUCCGCAAAAAAGGAG-A ..........(((.(((((((((....))))))))).)))....(((..(((((....(((......))))))))..)))..........-. ( -32.10) >DroSim_CAF1 42721 91 - 1 UAAAGAAGAUUCCAGUCAGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUGGCCGUUUCCAGCCAGUCCGCAAAAAAGGAG-A ..........(((.(((((((((....))))))))).)))....(((..(((((....(((......))))))))..)))..........-. ( -32.80) >DroEre_CAF1 39489 88 - 1 UAAAGAAGACUCCAGUCAGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUAGCCAUUUCAAGCCUGUCC--AAAAAAGGCG-- ....(((...(((.(((((((((....))))))))).)))....(((.((((......))))))))))..((((....--.....)))).-- ( -30.10) >DroYak_CAF1 41501 92 - 1 UAAAGAAGAUUCCAGUCAGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUAGCCAUUUCAAGCCAGUCCGCAAAAAAAGUGGA ..........(((.(((((((((....))))))))).)))....(((..(((((................)))))..)))............ ( -30.09) >consensus UAAAGAAGAUUCCAGUCAGCUUCUAAUGGAGCUGACCGGAUAACGUGUGCUGGUCCAGUGGCCGUUUCCAGCCAGUCCGCAAAAAAGGAG_A ..........(((.(((((((((....))))))))).)))....(((..(((((................)))))..)))............ (-27.37 = -27.81 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:18 2006