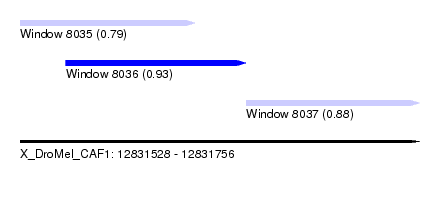
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,831,528 – 12,831,756 |
| Length | 228 |
| Max. P | 0.929442 |

| Location | 12,831,528 – 12,831,628 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -17.11 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12831528 100 + 22224390 AU--------------GCUAUAUUCCAAUAUCAUUCAUGUCAUAUCCUGGCGCCUGUAGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCU ..--------------................................((((.(((.(((..(((((((....)))))))((((((((....)))))))).)))------.)))..)))) ( -24.20) >DroSec_CAF1 29448 100 + 1 AU--------------GAUAUAUUCCAAUAUCAUUCAUGUCAUAUCCCGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCU ((--------------(((((......)))))))..............((((...(((((..(((((((....)))))))((((((((....)))))))).)))------))....)))) ( -27.40) >DroSim_CAF1 39219 100 + 1 AU--------------GCUAUAUUCCAAUAUCAUUCAUGCCAUAUCCCGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCU ..--------------((....................(((.......)))....(((((..(((((((....)))))))((((((((....)))))))).)))------)).....)). ( -26.00) >DroEre_CAF1 37188 86 + 1 AUGGU----------CGGUAUAUUCCAAUAUAAUUCAUGUCAUAUCCUGGCGCCCGAGGCAAACAU--------GCAUGUGAAAUUGAUGGAUCAGCUUCAGCU---------------- .((((----------(((......))........(((..(((((((.((..(((...)))...)).--------).))))))...)))..)))))((....)).---------------- ( -17.70) >DroYak_CAF1 38054 120 + 1 AUGGUAUAUGGUAUAUGGUGUAUUCCAAUAUCAUUCAUGUCAUAUCCUGGCGCCUGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUUUCAGUUUCAGUUCGGU (((..(((((((((.(((......))))))))))..))..)))...((((.((.((((((..(((((((....)))))))((((((((....)))))))).)))))))).))))...... ( -36.40) >consensus AU______________GCUAUAUUCCAAUAUCAUUCAUGUCAUAUCCUGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU______UCAGUUCGCU ......................................(((.......)))((.....))..(((((((....)))))))((((((((....)))))))).................... (-17.11 = -17.60 + 0.49)

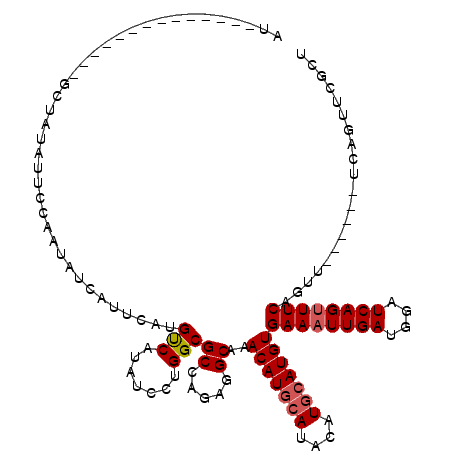

| Location | 12,831,554 – 12,831,657 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12831554 103 + 22224390 CAUAUCCUGGCGCCUGUAGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCUGGUAUAU----AUGUUUCUGUUUCUGCUUUUGU .((((.(..(((.(((.(((..(((((((....)))))))((((((((....)))))))).)))------.)))..)))..).))))----...................... ( -29.50) >DroSec_CAF1 29474 103 + 1 CAUAUCCCGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCUGGUAUAU----AUAUUUCUGUUUCUGCUUUUGU .((((.((((((...(((((..(((((((....)))))))((((((((....)))))))).)))------))....)))))).))))----...................... ( -31.40) >DroSim_CAF1 39245 103 + 1 CAUAUCCCGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU------UCAGUUCGCUGGUAUAU----AUGUUUCUGUUUCUGCUUUUGU .((((.((((((...(((((..(((((((....)))))))((((((((....)))))))).)))------))....)))))).))))----...................... ( -31.40) >DroYak_CAF1 38094 113 + 1 CAUAUCCUGGCGCCUGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUUUCAGUUUCAGUUCGGUGGUAUAUAUGUAUGUUUCUCUUUUUGCUUUUGU ........((((...((((.(((((((((((.((((.(((((((((((((((.....))))...)))))))))...))...)))).))))))))))).))))..))))..... ( -30.50) >consensus CAUAUCCCGGCGCCAGAGGCAAACAUGCAUACAUGCAUGUGAAAUUGAUGGAUCAGUUUCAGUU______UCAGUUCGCUGGUAUAU____AUGUUUCUGUUUCUGCUUUUGU .((((.(((((((.....))..(((((((....)))))))((((((((....)))))))).................))))).)))).......................... (-24.27 = -24.28 + 0.00)


| Location | 12,831,657 – 12,831,756 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.74 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12831657 99 + 22224390 UGUCGCCGCCUAUAUGGCUAUAUGAC--------------------UGGCUAUAUGGCUGUAUAG-UGCGCUUUUCUGGCCUCCGCCAAGUGGCUUCUGUUCUGAUAUUUGCAUAACGUU .(((((((((((((((((((((((..--------------------....)))))))))))))))-.)))......((((....)))).)))))....(((.((.......)).)))... ( -31.70) >DroSec_CAF1 29577 84 + 1 UGUCGCCGGCUA----------------------------------UGGCUAUAUGGCU--AUAGUUGCUCUUUUUUGGCCUCCGCCAAGUAGCUUCUGUUCUGAUAUUUGCAUAACGUU ((((...(((((----------------------------------(((((....))))--))))))(((....((((((....)))))).))).........))))............. ( -21.80) >DroSim_CAF1 39348 120 + 1 UGUCGCCGGCUAUAUGGAUGUAUGGCUAUAUAGCUGUAUGGCUAUAUGGCUAUAUGGCUGUAUAGUUGCUCUUUUCUGGCCUCCGCCAAGUGGCUUCUGUUCUGAUAUUUGCAUAACGUU .((((((((((((((((..((((((((((((((((....))))))))))))))))..)))))))))))........((((....)))).)))))....(((.((.......)).)))... ( -44.60) >DroEre_CAF1 37314 95 + 1 UGUCGCUGGCCAUGUGGC------------------------UAUAUGGAUAUAUGGCAAUAUAGUUGCUCUUUUCUGGCCUCCAUCAAGUGGCUUCUGCGCUGAUAUUUGCAUAACGU- .(((((((((((.(..((------------------------(((((............)))))))..).......))))).......))))))......((........)).......- ( -22.71) >DroYak_CAF1 38207 88 + 1 UGUCGCUGGC--------------------------------UAUAUGGCUGUAUGGCUGUAUAGUUGCUCUUUUCUGGCCUCCAUCAAGUGGCUUCUUCUCUGAUAUUUGCAUAACGUU ((((((.(((--------------------------------(((((((((....))))))))))))))........((((.(......).))))........))))............. ( -21.60) >consensus UGUCGCCGGCUAU_UGG_________________________UAUAUGGCUAUAUGGCUGUAUAGUUGCUCUUUUCUGGCCUCCGCCAAGUGGCUUCUGUUCUGAUAUUUGCAUAACGUU .(((((.........................................((((((((....)))))))).........((((....)))).))))).......................... ( -9.86 = -10.74 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:16 2006