
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,823,983 – 12,824,143 |
| Length | 160 |
| Max. P | 0.981012 |

| Location | 12,823,983 – 12,824,103 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -27.24 |
| Energy contribution | -27.55 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12823983 120 + 22224390 UUUAAAGUUGAGCCAAUUUUCCGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUA ....((((((((..........(((.(((....))).)))......(((((..(((((((......))))))).)))))..))))(((((.(((......))).)))))......)))). ( -30.30) >DroSec_CAF1 22040 119 + 1 U-UAAAGUUGAGCCAAUUUUCCGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAAUUCAUCGGAAACGGAAUCACGUCUCCGACAUUUCGCUUA .-......(((((.........(((.(((....))).)))......(((((..(((((((......))))))).)))))......(((((.(((......))).)))))......))))) ( -29.40) >DroEre_CAF1 30351 119 + 1 U-UAAAGUUGAGCCAAUUUUCCGGUUAAUGUUUCUCUGCCAGUCCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCCCCGACAUUUCGCUUA .-......(((((.((...((.((....((((((............(((((..(((((((......))))))).))))).......((....))))))))....)).))...)).))))) ( -28.00) >DroYak_CAF1 30680 119 + 1 U-UAAAGUUGAGCCAAUUUUCCGGUUAAUGUUUACUUGCCAGUUGAGGUAACUUUGUUUCGAAUUGGCAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUA .-......(((((.........(((.(((......(((((((((((..((....))..)).)))))))))..))).)))......(((((.(((......))).)))))......))))) ( -30.00) >consensus U_UAAAGUUGAGCCAAUUUUCCGGUUAAUGUUUAUUUGCCAGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUA ....((((((((..........(((............)))......(((((..(((((((......))))))).)))))..))))(((((.(((......))).)))))......)))). (-27.24 = -27.55 + 0.31)

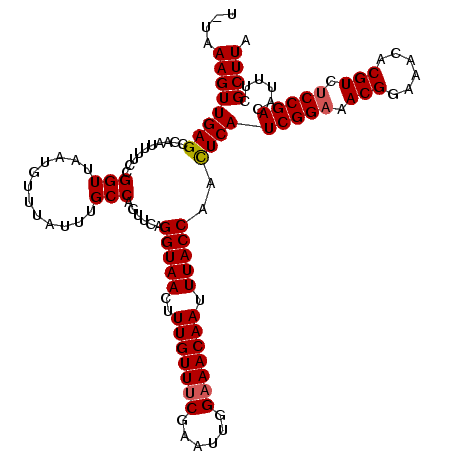

| Location | 12,823,983 – 12,824,103 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12823983 120 - 22224390 UAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCGGAAAAUUGGCUCAACUUUAAA ..........((((((((((......))))))))))((((((((((.(((((((......)))))))..)))))....((((..(((....)))..))))......)))))......... ( -33.30) >DroSec_CAF1 22040 119 - 1 UAAGCGAAAUGUCGGAGACGUGAUUCCGUUUCCGAUGAAUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCGGAAAAUUGGCUCAACUUUA-A ..(((.((..((((((((((......)))))))))).....(((((.(((((((......)))))))..)))))....((((..(((....)))..))))....)).)))........-. ( -31.20) >DroEre_CAF1 30351 119 - 1 UAAGCGAAAUGUCGGGGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGGACUGGCAGAGAAACAUUAACCGGAAAAUUGGCUCAACUUUA-A ..........(((((..(((......)))..)))))((((((((((.(((((((......)))))))..)))))....((((..((......))..))))......))))).......-. ( -32.80) >DroYak_CAF1 30680 119 - 1 UAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUGCCAAUUCGAAACAAAGUUACCUCAACUGGCAAGUAAACAUUAACCGGAAAAUUGGCUCAACUUUA-A ..........((((((((((......))))))))))((((((((((....)))))))))).....((((((.((....((((..(((....)))..))))......))...)))))).-. ( -32.10) >consensus UAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACUGGCAAAUAAACAUUAACCGGAAAAUUGGCUCAACUUUA_A ..........((((((((((......))))))))))((((((((((.(((((((......)))))))..)))))....((((..(((....)))..))))......)))))......... (-30.64 = -30.82 + 0.19)



| Location | 12,824,023 – 12,824,143 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -30.80 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12824023 120 + 22224390 AGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUAGAUAGUUUUGACUUGUCUGCGAAGUUCAGUUUCAAAUUGC ......(((((..(((((((......))))))).)))))((((..(((((.(((......))).))))).(((((((..(((((((....)).))))))))))))..))))......... ( -33.50) >DroSec_CAF1 22079 120 + 1 AGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAAUUCAUCGGAAACGGAAUCACGUCUCCGACAUUUCGCUUAGAUAGUUUUGAGUUGUCUGCGAAGUUCAGUUUAAAAUUGC ......(((((..(((((((......))))))).)))))......(((((.(((......))).))))).(((((((..((((((.......)))))))))))))............... ( -32.80) >DroEre_CAF1 30390 120 + 1 AGUCCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCCCCGACAUUUCGCUUAGAUAGUUUUGACUUGUCUGCGAAGUUAAGUUUCAAAUUGC ......(((((..(((((((......))))))).))))).......((....))(((((..(((...)))(((((((..(((((((....)).))))))))))))...)))))....... ( -32.30) >DroYak_CAF1 30719 120 + 1 AGUUGAGGUAACUUUGUUUCGAAUUGGCAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUAGAUAGUUUCGACUUGUCUGCGAAGUUCAGAUUCCAAUUGC (((((.((..((..((((((((((((....))))))..........((....)))))))).))..))((.(((((((..(((((((....)).))))))))))))))......))))).. ( -32.60) >consensus AGUUCAGGUAACUUUGUUUCGAAUUGGAAACAAUUUACCAACUCAUCGGAAACGGAAACACGUCUCCGACAUUUCGCUUAGAUAGUUUUGACUUGUCUGCGAAGUUCAGUUUCAAAUUGC ......(((((..(((((((......))))))).)))))......(((((.(((......))).))))).(((((((..((((((.......)))))))))))))............... (-30.80 = -31.30 + 0.50)



| Location | 12,824,023 – 12,824,143 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -28.64 |
| Energy contribution | -28.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12824023 120 - 22224390 GCAAUUUGAAACUGAACUUCGCAGACAAGUCAAAACUAUCUAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACU .........((((....((((((((..(((....))).)))..)))))..((((((((((......)))))))))).))))(((((.(((((((......)))))))..)))))...... ( -32.80) >DroSec_CAF1 22079 120 - 1 GCAAUUUUAAACUGAACUUCGCAGACAACUCAAAACUAUCUAAGCGAAAUGUCGGAGACGUGAUUCCGUUUCCGAUGAAUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACU .................((((((((.............)))..)))))..((((((((((......)))))))))).....(((((.(((((((......)))))))..)))))...... ( -29.12) >DroEre_CAF1 30390 120 - 1 GCAAUUUGAAACUUAACUUCGCAGACAAGUCAAAACUAUCUAAGCGAAAUGUCGGGGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGGACU .........(((((...((((((((..(((....))).)))..)))))..(((((..(((......)))..))))))))))(((((.(((((((......)))))))..)))))...... ( -33.10) >DroYak_CAF1 30719 120 - 1 GCAAUUGGAAUCUGAACUUCGCAGACAAGUCGAAACUAUCUAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUGCCAAUUCGAAACAAAGUUACCUCAACU ......((.........((((((((..(((....))).)))..)))))..((((((((((......))))))))))((((((((((....))))))))))............))...... ( -33.00) >consensus GCAAUUUGAAACUGAACUUCGCAGACAAGUCAAAACUAUCUAAGCGAAAUGUCGGAGACGUGUUUCCGUUUCCGAUGAGUUGGUAAAUUGUUUCCAAUUCGAAACAAAGUUACCUGAACU .................((((((((..(((....))).)))..)))))..((((((((((......)))))))))).....(((((.(((((((......)))))))..)))))...... (-28.64 = -28.95 + 0.31)

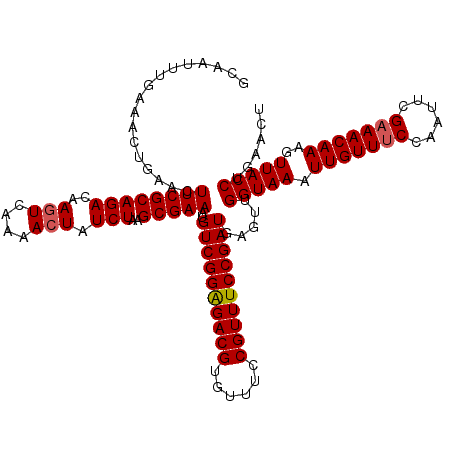
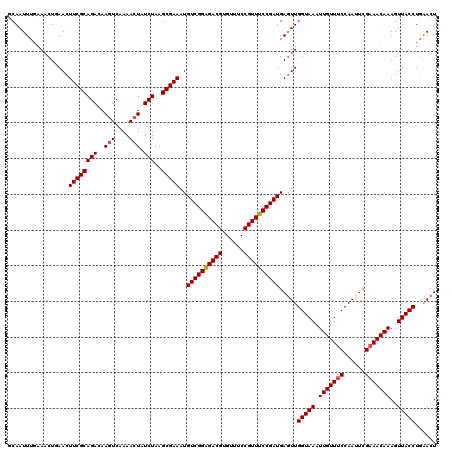
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:11 2006