
| Sequence ID | X_DroMel_CAF1 |
|---|---|
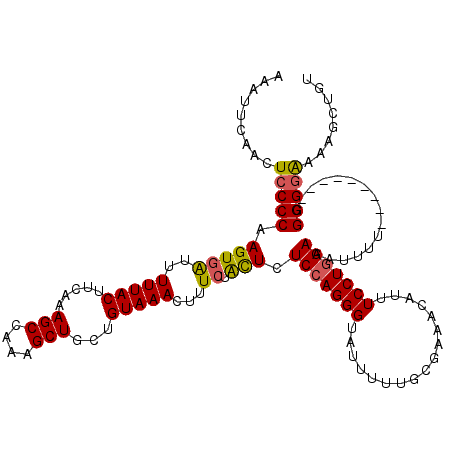
| Location | 12,823,796 – 12,823,907 |
| Length | 111 |
| Max. P | 0.893848 |

| Location | 12,823,796 – 12,823,907 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -20.97 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
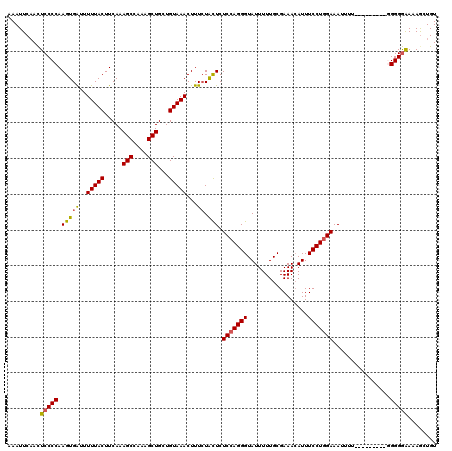
>X_DroMel_CAF1 12823796 111 + 22224390 AAAUUCAACUCCCCAAGUGAUUUUUACUUCAAAGCCAAAGCUGCUGUAAACUUUCUACUCUCCAGGGUAUUUUUGCGAAAAAUUUCCUGGAAAUUUU---------CGGGGAAAAGCUGU .........(((((.(((((..(((((.....(((....)))...)))))...)).))).(((((((.(((((.....))))).)))))))......---------.)))))........ ( -27.90) >DroSec_CAF1 21858 111 + 1 AAAUUCAAUUCCCCAAGUGAUUUUUACUUCAAAGCCAAAGCUGCUGUAAACUUUCUACUCUCCAGGGUAUUUUUGCGAAACAUUUCCUGGAAAUUUU---------GGGGAAAAAGCUGU ........((((((((((((..(((((.....(((....)))...)))))...)).))..(((((((.................)))))))....))---------))))))........ ( -27.13) >DroSim_CAF1 31611 111 + 1 AAAUUCAACUCCCCAAGUGAUUUUUACUUCAAAGCCAAAGCUGCUGUAAACUUUAUACUCUCCAGGGUAUUUUUGCGAAACAUUUCCUGGAAAUUUU---------GGGGGAAAAGCUGU .........(((((((((((..(((((.....(((....)))...)))))..))))....(((((((.................)))))))....))---------)))))......... ( -26.63) >DroEre_CAF1 30183 111 + 1 AAAUUCCACUCCCCAAGUGGUUUUUACUUCAAAGCCAAAGCUGAUGUAAACUUUCUAUUCUCCAGGGUAUUUUUGCGAAACAUUUCCUUGAAAUUUU---------GGGGGGAAAACUGU ........((((((((((((((((......)))))))(((..(((((........(((((....)))))..........)))))..))).....)))---------))))))........ ( -24.37) >DroYak_CAF1 30486 120 + 1 AAAUUCAACUCCCCAAGUGAUUUUUACUUCAAAGCCAAAGCUGAUGUAAACUUUCUGCUCUCCAGGGUAUUUUUGCGAAACAUUUCCUGGAAAUUUUUGUUUGGUUAGGGGAAUAGCUGU .........(((((((((((...))))))...(((((((((.((.........)).))..(((((((.................)))))))........))))))).)))))........ ( -28.53) >consensus AAAUUCAACUCCCCAAGUGAUUUUUACUUCAAAGCCAAAGCUGCUGUAAACUUUCUACUCUCCAGGGUAUUUUUGCGAAACAUUUCCUGGAAAUUUU_________GGGGGAAAAGCUGU .........(((((.(((((..(((((.....(((....)))...)))))...)).))).(((((((.................)))))))................)))))........ (-20.97 = -20.93 + -0.04)

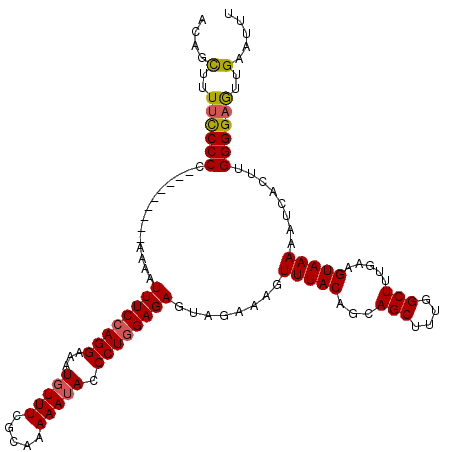
| Location | 12,823,796 – 12,823,907 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
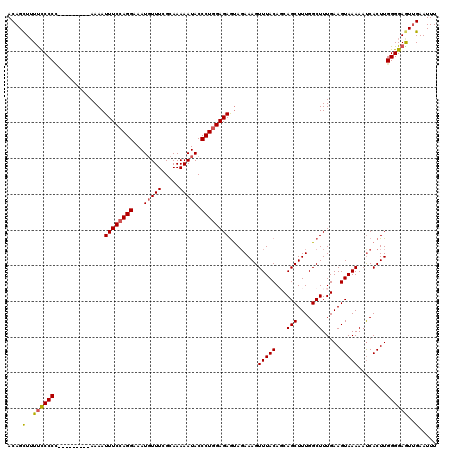
>X_DroMel_CAF1 12823796 111 - 22224390 ACAGCUUUUCCCCG---------AAAAUUUCCAGGAAAUUUUUCGCAAAAAUACCCUGGAGAGUAGAAAGUUUACAGCAGCUUUGGCUUUGAAGUAAAAAUCACUUGGGGAGUUGAAUUU .((((...((((((---------(...((((((((..(((((.....)))))..))))))))...((...(((((..((((....))..))..)))))..))..)))))))))))..... ( -31.30) >DroSec_CAF1 21858 111 - 1 ACAGCUUUUUCCCC---------AAAAUUUCCAGGAAAUGUUUCGCAAAAAUACCCUGGAGAGUAGAAAGUUUACAGCAGCUUUGGCUUUGAAGUAAAAAUCACUUGGGGAAUUGAAUUU ........((((((---------((..((((((((...(((((.....))))).))))))))...((...(((((..((((....))..))..)))))..))..))))))))........ ( -28.40) >DroSim_CAF1 31611 111 - 1 ACAGCUUUUCCCCC---------AAAAUUUCCAGGAAAUGUUUCGCAAAAAUACCCUGGAGAGUAUAAAGUUUACAGCAGCUUUGGCUUUGAAGUAAAAAUCACUUGGGGAGUUGAAUUU .(((((....((((---------((..((((((((...(((((.....))))).))))))))........(((((..((((....))..))..)))))......)))))))))))..... ( -28.40) >DroEre_CAF1 30183 111 - 1 ACAGUUUUCCCCCC---------AAAAUUUCAAGGAAAUGUUUCGCAAAAAUACCCUGGAGAAUAGAAAGUUUACAUCAGCUUUGGCUUUGAAGUAAAAACCACUUGGGGAGUGGAAUUU ..(((((..(((((---------((..((((.(((...(((((.....))))).))).))))........(((((.(((((....))..))).)))))......)))))).)..))))). ( -23.30) >DroYak_CAF1 30486 120 - 1 ACAGCUAUUCCCCUAACCAAACAAAAAUUUCCAGGAAAUGUUUCGCAAAAAUACCCUGGAGAGCAGAAAGUUUACAUCAGCUUUGGCUUUGAAGUAAAAAUCACUUGGGGAGUUGAAUUU .((((...(((((..............((((((((...(((((.....))))).))))))))...((...(((((.(((((....))..))).)))))..))....)))))))))..... ( -29.80) >consensus ACAGCUUUUCCCCC_________AAAAUUUCCAGGAAAUGUUUCGCAAAAAUACCCUGGAGAGUAGAAAGUUUACAGCAGCUUUGGCUUUGAAGUAAAAAUCACUUGGGGAGUUGAAUUU ....(..((((((..............((((((((...(((((.....))))).))))))))........(((((...(((....))).....)))))........))))))..)..... (-21.70 = -21.82 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:08 2006