| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,814,461 – 12,814,561 |
| Length | 100 |
| Max. P | 0.625692 |

| Location | 12,814,461 – 12,814,561 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.15 |
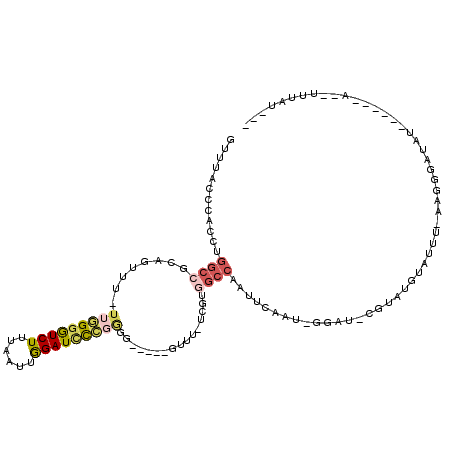
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -10.05 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625692 |
| Prediction | RNA |
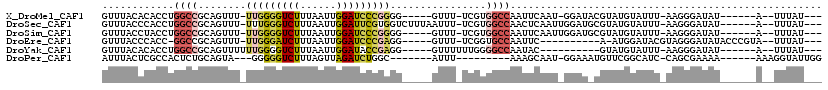
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12814461 100 - 22224390 GUUUACACACCUGGCCGCAGUUU-UUGGGGUCUUUAAUUGGAUCCCGGGG-----GUUU-UCGUGGCCAAUUCAAU-GGAUACGUAUGUAUUU-AAGGGAUAU------A--UUUAU--- ...........(((((((((..(-(((((((((......)))))))))).-----.)).-..)))))))((((..(-((((((....))))))-)..))))..------.--.....--- ( -32.50) >DroSec_CAF1 12513 106 - 1 GUUUACCCACCUGGCCGCAGUUU-UUUGGGUCUUUAAUUGGAUUCGUGGUCUUUAAUUU-UCGUGGCCAACUCAAUUGGAUGCGUAUGUAUUU-AAGGGAUAU------A--UUUAU--- .....(((...(((((((.....-...((((((......))))))..((..........-)))))))))......((((((((....))))))-)))))....------.--.....--- ( -26.50) >DroSim_CAF1 22899 101 - 1 GUUUACCUACCUGGCCGCAGUUU-UUGGGGUCUUUAAUUGGAUCCCGGGG-----GUUU-UCGUGGCCAAUUCAAUUGGAUGCGUAUGUAUUU-AAGGGAUAU------A--UUUAU--- .....(((...(((((((((..(-(((((((((......)))))))))).-----.)).-..)))))))......((((((((....))))))-)))))....------.--.....--- ( -33.50) >DroEre_CAF1 20698 96 - 1 GUUUACCCACC-GGCCGCAGUUU-UUGGGAUCUUUAAUUGGAUCCCGAGG-----GUUU-UCGGUGCCAAUUC----------A-AUGGAUACGUAGGGAUAUACCCGUA--UUUAU--- .......((((-((..((....(-(((((((((......)))))))))).-----))..-)))))).......----------.-(((((((((..((......))))))--)))))--- ( -30.80) >DroYak_CAF1 21944 93 - 1 GUUUACACACCUGGCCGCAGUUUUUUGGGGUCUUUAAUUGGAUACCGAGG-----GUUUUUUGGGGCCAAUAC----------GUAUGUAUUU-AAGGGAUAU------A--UUUAU--- .........(((((((.(((.(((((((..(((......)))..))))))-----)....))).))))(((((----------....))))).-.))).....------.--.....--- ( -24.80) >DroPer_CAF1 18373 93 - 1 AUUUACUCGCCACUCUGCAGUA---GGGGGUCUUUAGUUAGAUCUGGC-------AUUU---------AAAGCAAU-GGAAAUGUUCGGCAUC-CAGCGAAAA------AAAGGUAUUGG ...((((.(((((((((...))---)))(((((......)))))))))-------....---------...((..(-(((..((.....))))-)))).....------...)))).... ( -18.50) >consensus GUUUACCCACCUGGCCGCAGUUU_UUGGGGUCUUUAAUUGGAUCCCGGGG_____GUUU_UCGUGGCCAAUUCAAU_GGAU_CGUAUGUAUUU_AAGGGAUAU______A__UUUAU___ ............((((........(((((((((......)))))))))................)))).................................................... (-10.05 = -10.80 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:05 2006