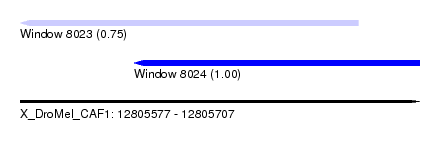
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,805,577 – 12,805,707 |
| Length | 130 |
| Max. P | 0.999766 |

| Location | 12,805,577 – 12,805,687 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.38 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -25.08 |
| Energy contribution | -27.20 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12805577 110 - 22224390 -------UCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUUUU -------.(((((.(((((.(((((.(((((((...))))))).)))))))))))).)))....((..(((((...((.((((((((....)))))))).))..)))))...))... ( -37.10) >DroSec_CAF1 3590 110 - 1 -------UCCUUGAGAUCCAUAAGGUCCUUGUGAUUCAUAAGGUCCUUGGGAUCCAUGGGCUCCUCGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUUUU -------(((.((.(((((.(((((.(((((((...))))))).)))))))))))).)))........(((((...((.((((((((....)))))))).))..)))))........ ( -37.00) >DroEre_CAF1 11401 108 - 1 UC---------UGAGAUCCGUAAGGUCCUUGAGACCCGUAAGGUCCUUGGGAUCCAUAGGCUCUUCCAGUUUCGAUGCUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUUUU ..---------((.(((((.(((((.(((((.......))))).)))))))))))).(((((....((((..(.((((..(((((((....))))))))))).)..)))).))))). ( -32.50) >DroYak_CAF1 13509 117 - 1 CUUGAGAUCCUUGAGAUCCAUAAGGUCCUUGAGAGCCGUAAGGUCCUUGGGCGCCAUAGGGUCCACGAGCUUGGAUGUUCGGUUGGUAGGGAUUAAUCGCAUAGGGAUUGUAGUUCU ((...((((((((.(((((.((.(((.((..((..((....))..))..)).))).)))))))...((((......))))(((((((....)))))))...))))))))..)).... ( -39.80) >consensus _______UCCUUGAGAUCCAUAAGGUCCUUGAGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAACGUUCGGUUGGUAAGGAUUAAUCGCAUAGGGAUUGUAGUUUU ........(((((.(((((.(((((.(((((((...))))))).)))))))))))).)))........(((((......((((((((....)))))))).....)))))........ (-25.08 = -27.20 + 2.12)



| Location | 12,805,614 – 12,805,707 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.01 |
| Mean single sequence MFE | -42.28 |
| Consensus MFE | -22.99 |
| Energy contribution | -25.05 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.54 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12805614 93 - 22224390 GGAGAUUGCGGCCCAUAGGG---------------------------UCCUUGAGAUCCAUAAGGUCCUUGUGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAACGUUC (((..(((.(((((...)))---------------------------.(((((.(((((.(((((.(((((((...))))))).)))))))))))).)))..)).)))..)))....... ( -36.30) >DroSec_CAF1 3627 93 - 1 GGAGAUUGCGGCCCAUAAGG---------------------------UCCUUGAGAUCCAUAAGGUCCUUGUGAUUCAUAAGGUCCUUGGGAUCCAUGGGCUCCUCGAGAUCCAACGUUC (((..(((.((.......((---------------------------(((.((.(((((.(((((.(((((((...))))))).)))))))))))).))))))).)))..)))....... ( -38.51) >DroEre_CAF1 11438 111 - 1 GGAGAUUGCGGCCUAUAUGAACCGUAGGCUUCAGGGGAACUC---------UGAGAUCCGUAAGGUCCUUGAGACCCGUAAGGUCCUUGGGAUCCAUAGGCUCUUCCAGUUUCGAUGCUC .(((((((.(((((((.......)))))))...(((((.((.---------((.(((((.(((((.(((((.......))))).)))))))))))).))..))))))))))))....... ( -39.40) >DroYak_CAF1 13546 120 - 1 GGAGAUUGUGGCCCAUAUGGACCGUAGGCUCCAGGAGAUCCUUGAGAUCCUUGAGAUCCAUAAGGUCCUUGAGAGCCGUAAGGUCCUUGGGCGCCAUAGGGUCCACGAGCUUGGAUGUUC (((..(((((((((....(((((...(((((((((.((.(((((.((((.....))))..))))))))))).)))))....)))))...)).)))))))..)))..((((......)))) ( -54.90) >consensus GGAGAUUGCGGCCCAUAUGG___________________________UCCUUGAGAUCCAUAAGGUCCUUGAGAUCCAUAAGGUCCUUGGGAUCCAUAGGCUCCACGAGAUCCAACGUUC (((..(((.(((((((.................((....))...........(.(((((.(((((.(((((((...))))))).))))))))))))))))))...)))..)))....... (-22.99 = -25.05 + 2.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:04 2006