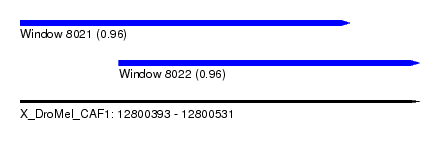
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,800,393 – 12,800,531 |
| Length | 138 |
| Max. P | 0.963142 |

| Location | 12,800,393 – 12,800,507 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12800393 114 + 22224390 -CCUUAAGCCUGGCAGUAACAAAUACUUGUGCAUAAACACAAACACACACACACACACGUGUUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACC -..((((...(((((((........(((((((((..((((((((((............))))))))))))).))))))(.(((......))))))))))).)))).......... ( -33.40) >DroSim_CAF1 8340 112 + 1 -CGGUAAGCCAGGCAGUAACAAAUACUCGUGCAUAAACACA--CAUACACACAUACACGUGGUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACC -.((....)).((((((...........((((.....((((--((((..(((......)))..)))))))).....(((((...)))))))))))))))................ ( -30.70) >DroEre_CAF1 6636 104 + 1 -ACUUAAGCCUGGCAGCAACAAAUACAUGUGCA----------CGAGCAUACACACAUGCGAGUAUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACC -..((((...((((((...........((((((----------..(((((.((((((((....)))))))).(....).)))))....)))))))))))).)))).......... ( -26.70) >DroYak_CAF1 7706 100 + 1 GCCUUAAGCCUGGUAGUAACAAAA-CA----AA----------CAGAUACACAUACAUUCAUAUGUGUGUGACACAAGGAUGGUAUUCAGCGCAUUGCUAAUUGAAUCAGAAACC .((((....(((...((......)-).----..----------)))...(((((((((....)))))))))....)))).(((((((.(((.....))))))...))))...... ( -18.40) >consensus _CCUUAAGCCUGGCAGUAACAAAUACAUGUGCA__________CAAACACACACACACGCGUUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACC ..........(((((((..................................((((((((....)))))))).....(((((...)))))....)))))))............... (-18.15 = -17.90 + -0.25)

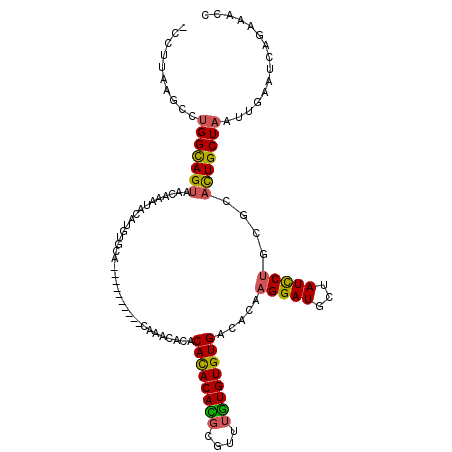
| Location | 12,800,427 – 12,800,531 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -19.42 |
| Energy contribution | -18.93 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12800427 104 + 22224390 AACACAAACACACACACACACACGUGUUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACCGCACCAUGAAAUCAGCAGGCAAAC ........((((((((.(((....))).)))))))).....(((((...)))))..((.(((((.(((...(((............)))))).))))))).... ( -27.90) >DroSim_CAF1 8374 102 + 1 AACACA--CAUACACACAUACACGUGGUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACCGCACCAUGAAAUCAGCAGGCAAAC ..((((--((((..(((......)))..)))))))).....(((((...)))))..((.(((((.(((...(((............)))))).))))))).... ( -27.10) >DroEre_CAF1 6668 96 + 1 --------CGAGCAUACACACAUGCGAGUAUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACCGCACCAUGAAAUCAGCAGGCAAAC --------...((...((((((((....)))))))).....(((((...)))))))((.(((((.(((...(((............)))))).))))))).... ( -25.50) >DroYak_CAF1 7734 95 + 1 --------CAGAUACACAUACAUUCAUAUGUGUGUGACACAAGGAUGGUAUUCAGCGCAUUGCUAAUUGAAUCAGAAACCGCACCAUAAAAUC-GCAAGCAAAC --------..(((.(((((((((....)))))))))......((.((((((((((.((...))...)))))).....))))..)).....)))-.......... ( -17.90) >consensus ________CAAACACACACACACGCGUUUGUGUGUGACACAAGGAUGCUAUCCUGCGCACUGCUAAUUGAAUCAGAAACCGCACCAUGAAAUCAGCAGGCAAAC ................((((((((....)))))))).....(((((...)))))..((.(((((.(((.....................))).))))))).... (-19.42 = -18.93 + -0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:02 2006