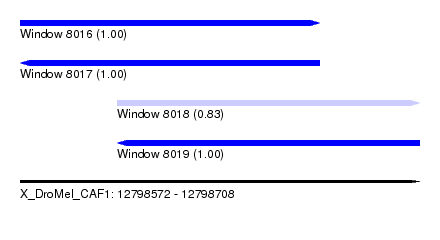
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,798,572 – 12,798,708 |
| Length | 136 |
| Max. P | 0.999983 |

| Location | 12,798,572 – 12,798,674 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.88 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.47 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.56 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12798572 102 + 22224390 AAACAAACACAGUGGCCAAACGCAGU---UUCACU--GCCCAUGAAAAUGGAAUCCAAUUUCCAUGU-----ACUUUUUUUUU----UUGCCACUGCCACUGCCAUUUCCGUGUUU ....((((((((((((.(((.((((.---....))--))........((((((......))))))..-----.........))----).))))))((....)).......)))))) ( -25.80) >DroSim_CAF1 4857 101 + 1 AAACAAACACAGUGGCCAAGCGCAGU---UUCACU--GCCCAUGAAAAUGGAAUCCAAUUUCCAUGU------CUUUUUUUUU----UGGCCACUGCCACUGCCAUUUCCGUGUUU ....((((((((((((((((.((((.---....))--))........((((((......))))))..------........))----))))))))((....)).......)))))) ( -32.00) >DroYak_CAF1 5640 94 + 1 AA----ACACAGUGGCCAAACGCAGU---UUCACU--GCCCAUGAAAAUGGAAUCCAAUUUCCAUGU-----ACUUUUUUUUUUUUUCGGCCACUGCCACUGCCA--------CUU ..----...((((((((....((((.---....))--))........((((((......))))))..-----................)))))))).........--------... ( -27.80) >DroAna_CAF1 5191 108 + 1 AA------GCAGUGU-CAGAAGCAGUAGAGACACUUGGGCAAUG-AAAUGGAAUCCAAUUUCCAUCUUAUAUACGUAUUUUUUUUUGUAUAUACAUACAUAUCUAGUUUGGUGUUU ((------(((((((-(............)))))(..(((....-..((((((......))))))..((((((((..........))))))))............)))..)))))) ( -19.90) >consensus AA____ACACAGUGGCCAAACGCAGU___UUCACU__GCCCAUGAAAAUGGAAUCCAAUUUCCAUGU_____ACUUUUUUUUU____UGGCCACUGCCACUGCCAUUUCCGUGUUU .........((((((((......(((......)))............((((((......)))))).......................)))))))).................... (-14.79 = -15.47 + 0.69)



| Location | 12,798,572 – 12,798,674 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.88 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.59 |
| SVM decision value | 5.32 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12798572 102 - 22224390 AAACACGGAAAUGGCAGUGGCAGUGGCAA----AAAAAAAAAGU-----ACAUGGAAAUUGGAUUCCAUUUUCAUGGGC--AGUGAA---ACUGCGUUUGGCCACUGUGUUUGUUU ....(((((....((((((((...(((..----...........-----.(((((((((.((...))))))))))).((--((....---.)))))))..)))))))).))))).. ( -32.90) >DroSim_CAF1 4857 101 - 1 AAACACGGAAAUGGCAGUGGCAGUGGCCA----AAAAAAAAAG------ACAUGGAAAUUGGAUUCCAUUUUCAUGGGC--AGUGAA---ACUGCGCUUGGCCACUGUGUUUGUUU ............(((((..((((((((((----(.........------.(((((((((.((...))))))))))).((--((....---.))))..)))))))))))..))))). ( -37.60) >DroYak_CAF1 5640 94 - 1 AAG--------UGGCAGUGGCAGUGGCCGAAAAAAAAAAAAAGU-----ACAUGGAAAUUGGAUUCCAUUUUCAUGGGC--AGUGAA---ACUGCGUUUGGCCACUGUGU----UU ...--------........((((((((((((.............-----.(((((((((.((...))))))))))).((--((....---.)))).))))))))))))..----.. ( -35.40) >DroAna_CAF1 5191 108 - 1 AAACACCAAACUAGAUAUGUAUGUAUAUACAAAAAAAAAUACGUAUAUAAGAUGGAAAUUGGAUUCCAUUU-CAUUGCCCAAGUGUCUCUACUGCUUCUG-ACACUGC------UU ...............(((((((((((............)))))))))))((((((((......))))))))-....((...((((((............)-)))))))------.. ( -21.60) >consensus AAACACGGAAAUGGCAGUGGCAGUGGCCA____AAAAAAAAAGU_____ACAUGGAAAUUGGAUUCCAUUUUCAUGGGC__AGUGAA___ACUGCGUUUGGCCACUGUGU____UU .............((((((((...(((.......................(((((((((.((...))))))))))).....(((......)))..)))..))))))))........ (-18.86 = -19.18 + 0.31)



| Location | 12,798,605 – 12,798,708 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.46 |
| Mean single sequence MFE | -12.80 |
| Consensus MFE | -6.38 |
| Energy contribution | -6.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12798605 103 + 22224390 CCCAUGAAAAUGGAAUCCAAUUUCCAUGU-----ACUUUUUUUUU----UUGCCACUGCCACUGCCAUUUCCGUGUUU---UUUCUCGCCA-UUCUUUCUUA--UAUCUAUUUUUUUU .....((((((((((......))))))..-----...........----.......................(((...---.....)))..-...))))...--.............. ( -8.10) >DroSim_CAF1 4890 97 + 1 CCCAUGAAAAUGGAAUCCAAUUUCCAUGU------CUUUUUUUUU----UGGCCACUGCCACUGCCAUUUCCGUGUUUU--UUUCUCGCCG-CUCUUUCUUA--UAUCUUUU------ ..((((...((((((......))))))..------..........----((((....))))..........))))....--..........-..........--........------ ( -12.80) >DroYak_CAF1 5669 94 + 1 CCCAUGAAAAUGGAAUCCAAUUUCCAUGU-----ACUUUUUUUUUUUUUCGGCCACUGCCACUGCCA--------CUU---UUCCGUGUUU-CUUUUUUCUUGAUAUCUUU------- ..((.((((((((((......))))))..-----................(((....))).....((--------(..---....)))...-....)))).))........------- ( -13.20) >DroAna_CAF1 5222 111 + 1 GCAAUG-AAAUGGAAUCCAAUUUCCAUCUUAUAUACGUAUUUUUUUUUGUAUAUACAUACAUAUCUAGUUUGGUGUUUUUUUUUUUUGCCAGCUUUGUUUUG--UAUUCUUU----UU ......-..((((((......))))))..((((((((..........)))))))).(((((.((..((((.((((...........))))))))..))..))--))).....----.. ( -17.10) >consensus CCCAUGAAAAUGGAAUCCAAUUUCCAUGU_____ACUUUUUUUUU____UGGCCACUGCCACUGCCAUUUCCGUGUUU___UUUCUCGCCA_CUCUUUCUUA__UAUCUUUU______ .........((((((......)))))).......................(((....))).......................................................... ( -6.38 = -6.12 + -0.25)

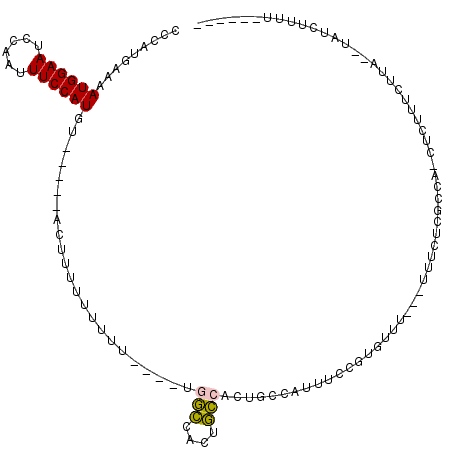
| Location | 12,798,605 – 12,798,708 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 66.46 |
| Mean single sequence MFE | -15.08 |
| Consensus MFE | -9.99 |
| Energy contribution | -9.05 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12798605 103 - 22224390 AAAAAAAAUAGAUA--UAAGAAAGAA-UGGCGAGAAA---AAACACGGAAAUGGCAGUGGCAGUGGCAA----AAAAAAAAAGU-----ACAUGGAAAUUGGAUUCCAUUUUCAUGGG ..............--..........-..........---...((.((((((((((((..(.(((....----...........-----.))).)..))))....)))))))).)).. ( -12.49) >DroSim_CAF1 4890 97 - 1 ------AAAAGAUA--UAAGAAAGAG-CGGCGAGAAA--AAAACACGGAAAUGGCAGUGGCAGUGGCCA----AAAAAAAAAG------ACAUGGAAAUUGGAUUCCAUUUUCAUGGG ------........--..........-..........--....((.((((((((.(((..((((..(((----..........------...)))..)))).))))))))))).)).. ( -14.82) >DroYak_CAF1 5669 94 - 1 -------AAAGAUAUCAAGAAAAAAG-AAACACGGAA---AAG--------UGGCAGUGGCAGUGGCCGAAAAAAAAAAAAAGU-----ACAUGGAAAUUGGAUUCCAUUUUCAUGGG -------.......((..........-...(((....---..)--------)).....(((....)))))..............-----.(((((((((.((...))))))))))).. ( -16.20) >DroAna_CAF1 5222 111 - 1 AA----AAAGAAUA--CAAAACAAAGCUGGCAAAAAAAAAAAACACCAAACUAGAUAUGUAUGUAUAUACAAAAAAAAAUACGUAUAUAAGAUGGAAAUUGGAUUCCAUUU-CAUUGC ..----........--.............((((......................(((((((((((............)))))))))))((((((((......))))))))-..)))) ( -16.80) >consensus ______AAAAGAUA__UAAAAAAAAG_UGGCAAGAAA___AAACACGGAAAUGGCAGUGGCAGUGGCCA____AAAAAAAAAGU_____ACAUGGAAAUUGGAUUCCAUUUUCAUGGG .........................................................((((....)))).....................(((((((((.((...))))))))))).. ( -9.99 = -9.05 + -0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:00 2006