
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,786,996 – 12,787,088 |
| Length | 92 |
| Max. P | 0.922872 |

| Location | 12,786,996 – 12,787,088 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.84 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
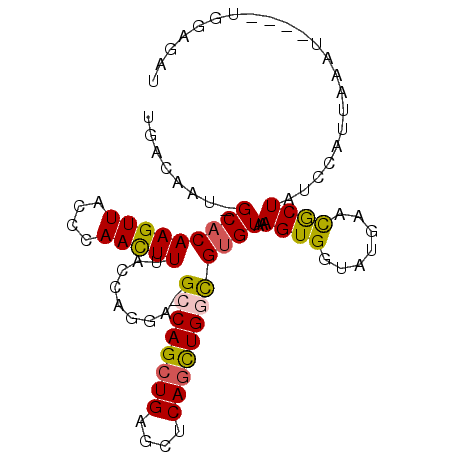
>X_DroMel_CAF1 12786996 92 + 22224390 UGACAAU-GCACAAGUUACCCAACUUUACCAGGA-GCCAGCUGAGCUCAGUUGGCGUGUAAAGUGAGAUGAACGCUAUCCAUUAAAU----UGGAGAU ......(-(((((((((....)))))........-((((((((....))))))))))))).((((.......)))).((((......----))))... ( -26.80) >DroSec_CAF1 1717 92 + 1 UGACAAU-GCACAAGUUACCCAACUUUACCAGGA-GCCAGCUGAGCUCAGCUGGCGUGUAAAGUGGUAUGAACGCUAUCCAUUAAAU----UGGAGAU .......-((......((((..(((((((.....-((((((((....))))))))..))))))))))).....))..((((......----))))... ( -31.10) >DroSim_CAF1 2757 93 + 1 UGACAAU-GCACAAGUUACUCAACUUUACCAAUACGCCAGCUGACCUCAUCUGGCGUGUAAAGUGGUAUGAACGCUAUCCAUUAAAU----UGGAUAU .......-((..(((((....)))))((((((((((((((.((....)).)))))))))....))))).....))((((((......----)))))). ( -28.80) >DroEre_CAF1 1622 90 + 1 UGACAAU-GCACAAGUUACCCAAUUUAACCAGGA-GGCAGCUGAGCUCAGCUGGUGUGUCGAGUGGGCUGAGCACUAUCCAUUAAAU----UGGAG-- .......-...........(((((((((...(((-....(((.((((((.((.(.....).)))))))).)))....))).))))))----)))..-- ( -27.70) >DroYak_CAF1 4034 95 + 1 UGACAAUGGCACAAGUUAGCCAACUUUAGCUAGA-GGCAGCUGAGCUCAGCUGGUGCGUAAAGUGGUAUGAAUACUAUCCAUUAAAUAACAUAAAA-- .....(((((((...(((((........))))).-..((((((....))))))))))....(((((.(((.....))))))))......)))....-- ( -24.00) >consensus UGACAAU_GCACAAGUUACCCAACUUUACCAGGA_GCCAGCUGAGCUCAGCUGGCGUGUAAAGUGGUAUGAACGCUAUCCAUUAAAU____UGGAGAU ........(((((((((....))))).........((((((((....))))))))))))..((((.......))))...................... (-18.00 = -17.84 + -0.16)

| Location | 12,786,996 – 12,787,088 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.18 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -17.06 |
| Energy contribution | -18.06 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
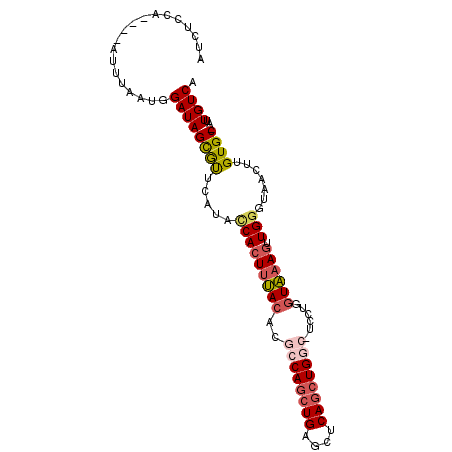
>X_DroMel_CAF1 12786996 92 - 22224390 AUCUCCA----AUUUAAUGGAUAGCGUUCAUCUCACUUUACACGCCAACUGAGCUCAGCUGGC-UCCUGGUAAAGUUGGGUAACUUGUGC-AUUGUCA .......----...(((((.((((.(((.((((.(((((((..((((.(((....))).))))-.....))))))).))))))))))).)-))))... ( -27.00) >DroSec_CAF1 1717 92 - 1 AUCUCCA----AUUUAAUGGAUAGCGUUCAUACCACUUUACACGCCAGCUGAGCUCAGCUGGC-UCCUGGUAAAGUUGGGUAACUUGUGC-AUUGUCA .......----...(((((.((((.(((....(((((((((..((((((((....))))))))-.....)))))).)))..))))))).)-))))... ( -31.80) >DroSim_CAF1 2757 93 - 1 AUAUCCA----AUUUAAUGGAUAGCGUUCAUACCACUUUACACGCCAGAUGAGGUCAGCUGGCGUAUUGGUAAAGUUGAGUAACUUGUGC-AUUGUCA .((((((----......))))))(((....((((((((((((((((((.((....)).)))))))....)))))).)).))).....)))-....... ( -27.90) >DroEre_CAF1 1622 90 - 1 --CUCCA----AUUUAAUGGAUAGUGCUCAGCCCACUCGACACACCAGCUGAGCUCAGCUGCC-UCCUGGUUAAAUUGGGUAACUUGUGC-AUUGUCA --.....----........((((((((.(((((((..........((((((....))))))((-....))......)))))....)).))-)))))). ( -28.10) >DroYak_CAF1 4034 95 - 1 --UUUUAUGUUAUUUAAUGGAUAGUAUUCAUACCACUUUACGCACCAGCUGAGCUCAGCUGCC-UCUAGCUAAAGUUGGCUAACUUGUGCCAUUGUCA --.................((((((.........((((((.((..((((((....))))))..-....)))))))).(((........))))))))). ( -22.20) >consensus AUCUCCA____AUUUAAUGGAUAGCGUUCAUACCACUUUACACGCCAGCUGAGCUCAGCUGGC_UCCUGGUAAAGUUGGGUAACUUGUGC_AUUGUCA ...................((((((((.....(((((((((..((((((((....))))))))......)))))).))).......))))...)))). (-17.06 = -18.06 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:52 2006