| Sequence ID | X_DroMel_CAF1 |
|---|---|
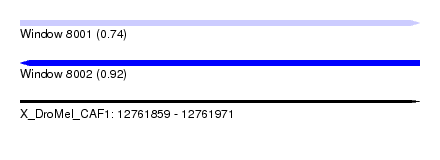
| Location | 12,761,859 – 12,761,971 |
| Length | 112 |
| Max. P | 0.922129 |

| Location | 12,761,859 – 12,761,971 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -22.11 |
| Energy contribution | -23.36 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
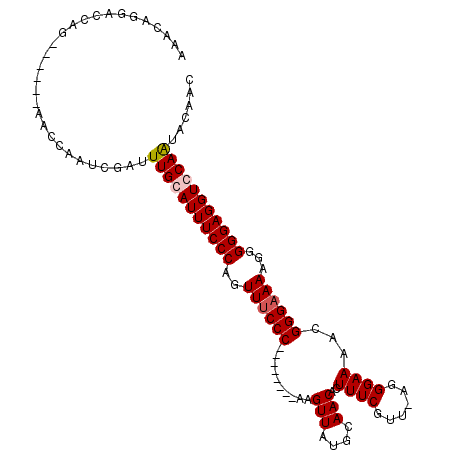
>X_DroMel_CAF1 12761859 112 + 22224390 GUUGUGUUGCACCUCCCCCUUUUCCCGUUUUCCCU-AACGAAAGUGUUGCAUAACUUGGCAGCUUGGGAAACUGGGAAAUGCAAAUCGAUUGGUUUUGGUUCUGGUCCUGUUU ......(((((.........(((((((.((((((.-.((....))(((((........)))))..)))))).))))))))))))...(((..(........)..)))...... ( -33.30) >DroSec_CAF1 16855 112 + 1 GUGGUAUUGCACCUCCCCCUUUUCCCGUUUUCCCA-AACGAAAGUGUUGCAUAACUUGGGAACUUGGGAAACUGGGAAAUGCAAAUCGAUUGGUUCUGGCUCUGGUCCUGUUU ..(((.(((((..((((...(((((((..((((((-(((....))(((....))))))))))..)))))))..))))..))))))))(((..(........)..)))...... ( -38.80) >DroEre_CAF1 18039 98 + 1 GUUGCACUGGACCUCCCCCUUUUCCCGUUUUCCCU-AACGAAAGUGUUGCAUAACUU--------GGGAAACGGGGAAAUGCAAAUCGAUUGGUU------CUGGUCCUGUUU .....((.(((((......((((((((((((((..-.((....))(((....)))..--------))))))))))))))........((.....)------).))))).)).. ( -30.20) >DroYak_CAF1 19411 99 + 1 GUUGUAUUGGACCUCCCCCUUUUCCCGUUUUCCCUUUACGAAAGUGUUGCAUAACUU--------GGGUAACAGGGAAAUGCAAAUCGAUUGGUU------CUGGUCCUGUUU ........(((((....((..((...(((((((((((((..((((........))))--------..)))).))))))).)).....))..))..------..)))))..... ( -25.30) >consensus GUUGUAUUGCACCUCCCCCUUUUCCCGUUUUCCCU_AACGAAAGUGUUGCAUAACUU________GGGAAACUGGGAAAUGCAAAUCGAUUGGUU______CUGGUCCUGUUU ........(((((.......(((((((.((((((.......((((........))))........)))))).)))))))..(((.....)))...........)))))..... (-22.11 = -23.36 + 1.25)



| Location | 12,761,859 – 12,761,971 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12761859 112 - 22224390 AAACAGGACCAGAACCAAAACCAAUCGAUUUGCAUUUCCCAGUUUCCCAAGCUGCCAAGUUAUGCAACACUUUCGUU-AGGGAAAACGGGAAAAGGGGGAGGUGCAACACAAC .....((.......)).............(((((((((((..((((((.((((....)))).........((((...-...))))..))))))...)))))))))))...... ( -30.30) >DroSec_CAF1 16855 112 - 1 AAACAGGACCAGAGCCAGAACCAAUCGAUUUGCAUUUCCCAGUUUCCCAAGUUCCCAAGUUAUGCAACACUUUCGUU-UGGGAAAACGGGAAAAGGGGGAGGUGCAAUACCAC .....((..........((.....))...(((((((((((..((((((...((((((((..(..........)..))-))))))...))))))...)))))))))))..)).. ( -37.00) >DroEre_CAF1 18039 98 - 1 AAACAGGACCAG------AACCAAUCGAUUUGCAUUUCCCCGUUUCCC--------AAGUUAUGCAACACUUUCGUU-AGGGAAAACGGGAAAAGGGGGAGGUCCAGUGCAAC .....(((((..------...(((.....)))...((((((.((((((--------..(((....)))..((((...-...))))..)))))).)))))))))))........ ( -30.40) >DroYak_CAF1 19411 99 - 1 AAACAGGACCAG------AACCAAUCGAUUUGCAUUUCCCUGUUACCC--------AAGUUAUGCAACACUUUCGUAAAGGGAAAACGGGAAAAGGGGGAGGUCCAAUACAAC .....(((((..------...(((.....)))..(..((((.((.(((--------..(((....)))..(..(.....)..)....))))).))))..))))))........ ( -24.40) >consensus AAACAGGACCAG______AACCAAUCGAUUUGCAUUUCCCAGUUUCCC________AAGUUAUGCAACACUUUCGUU_AGGGAAAACGGGAAAAGGGGGAGGUCCAAUACAAC .............................(((((((((((..((((((..........(((....)))..((((......))))...))))))...)))))))))))...... (-22.06 = -22.62 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:45 2006