| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,737,080 – 12,737,185 |
| Length | 105 |
| Max. P | 0.783838 |

| Location | 12,737,080 – 12,737,185 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -18.25 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
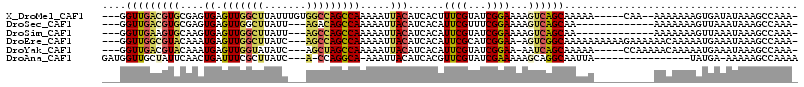
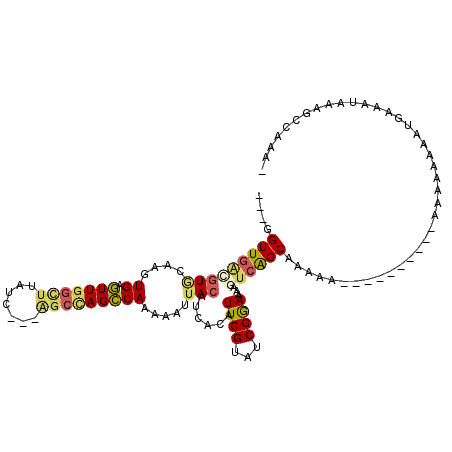
>X_DroMel_CAF1 12737080 105 + 22224390 ---GGUUGACGUGCGAGUGAGUUGGCUUAUUUGUGGCCAGCCAAAAAUUACAUCACUUUCGUAUCGGAAAAGUCAGCAAAAA-----CAA--AAAAAAAGUGAUAUAAAGCCAAA- ---.(((((((((((((((((((((((.......)))))))...........))))..)))))).......)))))).....-----...--.......................- ( -22.91) >DroSec_CAF1 1551 96 + 1 ---GGUUGACGUGCGAGUGAGUUGGCUUAUU---AGACAGCCAAAAAUUACAUCACAUUCGUUUCGGAAAAGUCAGCAA-------------AAAAAAAGUUAAAUAAAGCCAAA- ---.(((((((((...((((.((((((....---....))))))...))))..))).((((...))))...))))))..-------------.......................- ( -21.80) >DroSim_CAF1 546 96 + 1 ---GGUUGAAGUGCAAGUGAGUUGGCUUAUU---AGCCAGCCAAAAAUUACAUCACAUUCGUAUCGGAAAAGUCAGCAA-------------AAAAAAAGUUAAAUAAAGCCAAA- ---.(((((.(((...((((.((((((....---....))))))...))))..))).((((...))))....)))))..-------------.......................- ( -18.10) >DroEre_CAF1 1546 108 + 1 ---GGUUGGCGUACAAAUGAGUUGGCUUAUC---AGCCAGCCAAAAAUUACAUCACAUUCGCAUCGGAA-AGUCGGCAAAAAAAAAAGAAAAAACAAAAAUGAAAUAAAGCCAAA- ---..((((((((....((.(((((((....---))))))))).....))).........((..(....-.)...))................................))))).- ( -20.30) >DroYak_CAF1 1591 103 + 1 ---GGUUGACGUACAAAUGAGUUGGUAUAUC---AGCUAGCCAAAAAUUACAUCACAUUCGUAUCGGAA-AAUCAGCAAAAA-----CCAAAAACAAAAAUGAAAUAAAGCCAAA- ---.(((((.(((....((.((((((.....---.)))))))).....)))......((((...)))).-..))))).....-----............................- ( -11.70) >DroAna_CAF1 1435 94 + 1 GAUGGUUGCUAUUCAACUGAUUUCGCUUAUC---A-CCAGGCA-AAAUUACAUCACGUUCGUAUCGAAAAAGCAGGCAAUUA----------------UAUGA-AAAAAGCCAAAA .(((((((((.......((((((.((((...---.-..)))).-)))))).......((((...))))......))))))))----------------)....-............ ( -14.70) >consensus ___GGUUGACGUGCAAGUGAGUUGGCUUAUC___AGCCAGCCAAAAAUUACAUCACAUUCGUAUCGGAAAAGUCAGCAAAAA__________AAAAAAAAUGAAAUAAAGCCAAA_ ....(((((((((....((.(((((((.......))))))))).....)))......((((...))))...))))))....................................... (-11.69 = -12.03 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:40 2006