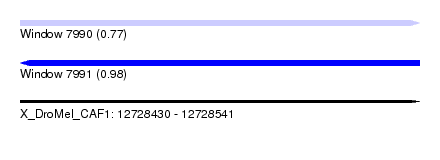
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,728,430 – 12,728,541 |
| Length | 111 |
| Max. P | 0.977750 |

| Location | 12,728,430 – 12,728,541 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.76 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12728430 111 + 22224390 AGCACAAAACAGAACCGCACAGAACAGCACCGACACAAAUGCAGAUACAAAUGUGGAUGUAUCUUCGCAACAGCAUCAGUAUCUGAAUCUGUAUCUGUAUCUGUAACUGUG ........((((....((........))...........((((((((((.(((..(((........((....)).((((...)))))))..))).)))))))))).)))). ( -28.30) >DroSec_CAF1 81095 105 + 1 AGCACAAAACAGAACCGCACAGAACAGCACCGACACAAAUGCAGAUACAAAUGUGGAUGUAUCUUCGCAGCAGCAGCAGUAUC------UGUAUCUGUAUCUGUAACUGUG ..((((..(((((...((........))......(((.((((((((((...((((((......))))))((....)).)))))------))))).))).)))))...)))) ( -30.70) >DroEre_CAF1 79919 88 + 1 AGCACAAAACAGAA-----CAGAACAGCACCGACACAAAUUCAGAUACAAAUGUGGAUGUAUCUUCGCAUCAGCUUCA------------GU------AUCUGUAACUGUG ..............-----..............((((....(((((((....((.(((((......))))).))....------------))------)))))....)))) ( -18.60) >DroYak_CAF1 84137 94 + 1 AGCACAAAACAGAG-----CAGAACAGCACCGACACAAAUGCAGAUACAAAUGUGGAUGUAUCUUCGCAUCAGCAUCU------------GUAUCUGUAUCUGUAACUGUG ........((((.(-----(......))......(((.(((((((((((.((((.(((((......))))).)))).)------------)))))))))).)))..)))). ( -32.20) >consensus AGCACAAAACAGAA_____CAGAACAGCACCGACACAAAUGCAGAUACAAAUGUGGAUGUAUCUUCGCAUCAGCAUCA____________GUAUCUGUAUCUGUAACUGUG .................................((((..((((((((((..((((((......))))))...((................))...))))))))))..)))) (-14.77 = -15.76 + 1.00)

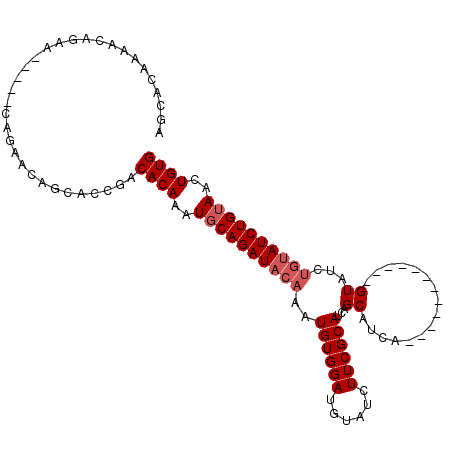

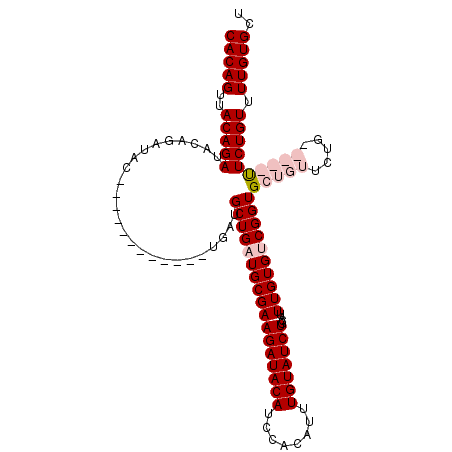
| Location | 12,728,430 – 12,728,541 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -21.39 |
| Energy contribution | -23.70 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12728430 111 - 22224390 CACAGUUACAGAUACAGAUACAGAUUCAGAUACUGAUGCUGUUGCGAAGAUACAUCCACAUUUGUAUCUGCAUUUGUGUCGGUGCUGUUCUGUGCGGUUCUGUUUUGUGCU (((((..(((((.((.(.((((((..(((.(((((((((...(((..(((((((........))))))))))...)))))))))))).))))))).))))))).))))).. ( -42.10) >DroSec_CAF1 81095 105 - 1 CACAGUUACAGAUACAGAUACA------GAUACUGCUGCUGCUGCGAAGAUACAUCCACAUUUGUAUCUGCAUUUGUGUCGGUGCUGUUCUGUGCGGUUCUGUUUUGUGCU (((((..(((((((((((.(((------(.(((((.(((...(((..(((((((........))))))))))...))).)))))))))))))))....))))).))))).. ( -35.60) >DroEre_CAF1 79919 88 - 1 CACAGUUACAGAU------AC------------UGAAGCUGAUGCGAAGAUACAUCCACAUUUGUAUCUGAAUUUGUGUCGGUGCUGUUCUG-----UUCUGUUUUGUGCU (((((..(((((.------((------------.((((((((((((((((((((........)))))))....))))))))))....))).)-----)))))).))))).. ( -27.90) >DroYak_CAF1 84137 94 - 1 CACAGUUACAGAUACAGAUAC------------AGAUGCUGAUGCGAAGAUACAUCCACAUUUGUAUCUGCAUUUGUGUCGGUGCUGUUCUG-----CUCUGUUUUGUGCU (((((..(((((..((((.((------------((.((((((((((((((((((........)))))))....)))))))))))))))))))-----.))))).))))).. ( -35.50) >consensus CACAGUUACAGAUACAGAUAC____________UGAUGCUGAUGCGAAGAUACAUCCACAUUUGUAUCUGCAUUUGUGUCGGUGCUGUUCUG_____UUCUGUUUUGUGCU (((((..(((((.........................(((((((((((((((((........)))))))....))))))))))(((((.....)))))))))).))))).. (-21.39 = -23.70 + 2.31)

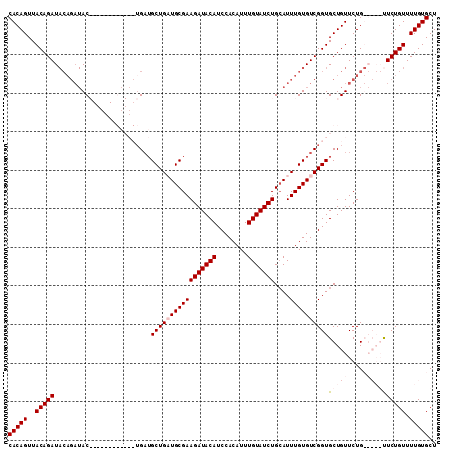
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:35 2006