
| Sequence ID | X_DroMel_CAF1 |
|---|---|
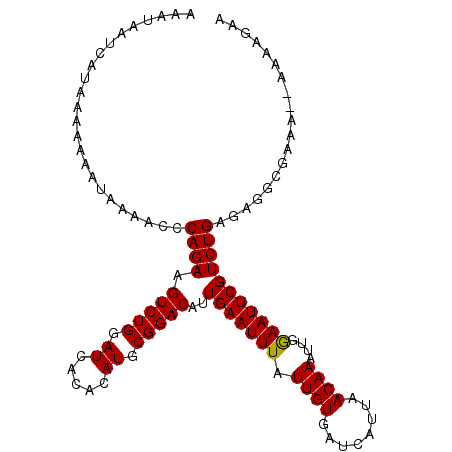
| Location | 12,722,687 – 12,722,817 |
| Length | 130 |
| Max. P | 0.958258 |

| Location | 12,722,687 – 12,722,795 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12722687 108 - 22224390 AAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACGUUGAAUUUAUUGUGAUCAUUAACAAAUUGGAAUUCGUCUGAGGGGCGAAA--AAAAGAA ......................((((((.(((((.((.....)).)))))...((((((.((((........))))....)))))).)))).)).......--....... ( -19.40) >DroSec_CAF1 74925 108 - 1 AAAUAAUCAUAAAAAAAAAAACACCAGAAGUCUGGAUCACACAUGCGGACAAUGAAUUUAUUGUGAUCAUUAACAAAUUGGAAUUCGUCUGAGAGGCGAAA--AAAAGAA ......((...............((((..(((((.((.....)).)))))..........((((........)))).))))..((((((.....)))))).--....)). ( -18.50) >DroSim_CAF1 60078 108 - 1 AAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACAAAUUGGAAUUCGUCUGAGAGGCGAAA--AAAAGAA ......((...............((((..(((((.((.....)).)))))..........((((........)))).))))..((((((.....)))))).--....)). ( -18.30) >DroEre_CAF1 74244 109 - 1 UAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACGUUGAAUUUAUUGUGAUCAUUAACAAAUUGAAAUUCGUCUGAGAGCCGAAA-AAAAAGAA ........................((((.(((((.((.....)).)))))...((((((.((((........))))....)))))).))))..........-........ ( -15.80) >DroYak_CAF1 78605 110 - 1 AAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACAAAUUGGAAUUCGUCUGAGAGCCAAAAAAAAAAGAA ........................((((.(((((.((.....)).)))))...((((((.((((........))))....)))))).))))................... ( -16.20) >consensus AAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACAAAUUGGAAUUCGUCUGAGAGGCGAAA__AAAAGAA ........................((((.(((((.((.....)).)))))..(((((((.((((........))))....)))))))))))................... (-16.30 = -16.14 + -0.16)


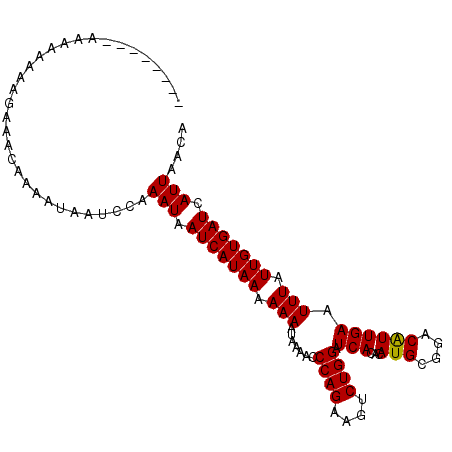
| Location | 12,722,720 – 12,722,817 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -11.45 |
| Consensus MFE | -10.82 |
| Energy contribution | -10.57 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12722720 97 - 22224390 --------AAAAAAGAGAAACA-AAUAAUCCAAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACGUUGAAUUUAUUGUGAUCAUUAACA --------..............-.........(((.(((((((.(((.......((((....)))).(((.((........))))).))).))))))).))).... ( -11.50) >DroSim_CAF1 60111 106 - 1 AAAACUAGAAAAAAAAGAAACAAAAUAAUCCAAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACA ................................(((.(((((((.(((.......((((....)))).(((...(((....)))))).))).))))))).))).... ( -10.80) >DroEre_CAF1 74278 99 - 1 -------AAAUAAAUAAACACAAAGUAAUCCUAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACGUUGAAUUUAUUGUGAUCAUUAACA -------........................((((.(((((((.(((.......((((....)))).(((.((........))))).))).))))))).))))... ( -12.70) >DroYak_CAF1 78640 98 - 1 --------CACACCCAUACACAAAAUAAUCCAAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACA --------........................(((.(((((((.(((.......((((....)))).(((...(((....)))))).))).))))))).))).... ( -10.80) >consensus ________AAAAAAAAGAAACAAAAUAAUCCAAAUAAUCAUAAAAAAAUAAAACCCAGAAGUCUGGAUCACACAUGCGGACAUUGAAUUUAUUGUGAUCAUUAACA ................................(((.(((((((.(((.......((((....)))).(((...(((....)))))).))).))))))).))).... (-10.82 = -10.57 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:32 2006