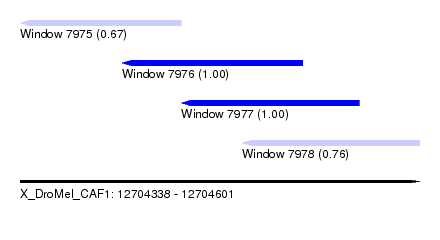
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,704,338 – 12,704,601 |
| Length | 263 |
| Max. P | 0.999315 |

| Location | 12,704,338 – 12,704,444 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -21.37 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12704338 106 - 22224390 CUUUUUCCGGCCAUCUGGUUG-GAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUUAGAUUUCGGCAAAACUUAUCUUUUUGCCAUUU------ ....((((((((....)))))-)))...(((..(((........)))..)))........((((...(((....))).))))(((((((.......)))))))....------ ( -22.70) >DroSec_CAF1 57062 111 - 1 CUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUUAUCUU--UGCCAUUUUUAUAU ...(((((((((....)))))))))...(((..(((........)))..)))........((((((((....).))).))))((((((.......))--)))).......... ( -29.10) >DroSim_CAF1 43557 111 - 1 CUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUUAUCUU--UGCCAUUUUUAUAU ...(((((((((....)))))))))...(((..(((........)))..)))........((((((((....).))).))))((((((.......))--)))).......... ( -29.10) >consensus CUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAGUUUAAUGAUCCAUACACAACGAAGCUCCUAAAGUGAGAUUUCGGCAAAACUUAUCUU__UGCCAUUUUUAUAU ...(((((((((....)))))))))...(((..(((........)))..)))........(((((((.......))).))))((((.............)))).......... (-21.37 = -21.82 + 0.45)


| Location | 12,704,405 – 12,704,524 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -23.36 |
| Energy contribution | -27.05 |
| Covariance contribution | 3.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12704405 119 - 22224390 AGGAUAUUCUAUAGAAAUACUCCAAAUUACAUCGUGACAUCUGAUAUGAUUUUGUUUGGGAAAGUCCAUGCAGCAAUGCACUUUUUCCGGCCAUCUGGUUG-GAAUACUGGUCCAUAAAG .((((.(((..((....))..((((((...((((((.(....).))))))...))))))))).))))..(((....)))(((..((((((((....)))))-)))....)))........ ( -31.90) >DroSec_CAF1 57133 120 - 1 AGGACAUUCUAUAGAAAUACUCCAAAUGGCAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCAUGCAGCAAUGCACUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAG .((((.(((..((....))..(((((..((((((((.(....).))))))))..)))))))).)))).((((....))))...(((((((((....)))))))))............... ( -44.80) >DroSim_CAF1 43628 120 - 1 AGGACAUUCUAUAGAAAUACUCGAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCGUGCAGCAAUGCACUUUUUCCGGCCAUCUGGCUGGGAAUACUGGUCCAUAAAG .((((.(((....)))...(((.(((..((((((((.(....).))))))))..))))))...))))(((((....)))))..(((((((((....)))))))))............... ( -44.00) >DroEre_CAF1 56652 98 - 1 AGGACAUUCUAUAGGAAUACUCCAAAUGACAUCAUGACAUUUGAUAUCCUGUCUAU--------AUC--------AUGCAC-UUUUCCGGCCAUCUGGAUG-GAGUACUGGAAUGU---- ...((((((((......(((((((........(((((.((..(((.....))).))--------.))--------)))...-...(((((....)))))))-))))).))))))))---- ( -23.70) >consensus AGGACAUUCUAUAGAAAUACUCCAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCAUGCAGCAAUGCACUUUUUCCGGCCAUCUGGCUG_GAAUACUGGUCCAUAAAG .((((.(((....)))....((((((((((((((((.(....).))))))))))))))))...)))).((((....))))...(((((((((....)))))))))............... (-23.36 = -27.05 + 3.69)

| Location | 12,704,444 – 12,704,561 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -23.92 |
| Energy contribution | -26.55 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12704444 117 - 22224390 UCGGUAGAGCGG---UAUUUUUAAGUCUAGGAUGUCAGAAAGGAUAUUCUAUAGAAAUACUCCAAAUUACAUCGUGACAUCUGAUAUGAUUUUGUUUGGGAAAGUCCAUGCAGCAAUGCA ..((.....(((---((((((((....(((((((((......)))))))))))))))))))((((((...((((((.(....).))))))...))))))).....)).((((....)))) ( -30.90) >DroSec_CAF1 57173 115 - 1 UCGGUAGAGCGG---UAUUCU--AGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGGCAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCAUGCAGCAAUGCA ..((.....(((---((((((--(...((((((((((....))))))))))))).))))))(((((..((((((((.(....).))))))))..)))))).....)).((((....)))) ( -43.70) >DroSim_CAF1 43668 115 - 1 UCGGUAGAGCGG---UAUUCU--AUUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCGAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCGUGCAGCAAUGCA ........((((---..((((--(...((((((((((....)))))))))))))))...(((.(((..((((((((.(....).))))))))..)))))).....))))(((....))). ( -40.80) >DroEre_CAF1 56686 102 - 1 UCAGUAAAGGGGGGCUAUUCU--AGUCUAGAAUGUCUAAAAGGACAUUCUAUAGGAAUACUCCAAAUGACAUCAUGACAUUUGAUAUCCUGUCUAU--------AUC--------AUGCA ...((.....((((.((((((--....((((((((((....))))))))))..)))))))))).....))..(((((.((..(((.....))).))--------.))--------))).. ( -27.60) >consensus UCGGUAGAGCGG___UAUUCU__AGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGACAUCAUGACAUCUGAUAUGAUGUUGUUUGGGAAAGUCCAUGCAGCAAUGCA ...(((..((.................((((((((((....)))))))))).........((((((((((((((((.(....).))))))))))))))))............))..))). (-23.92 = -26.55 + 2.63)


| Location | 12,704,484 – 12,704,601 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12704484 117 - 22224390 UGAUUUCGAUUAAAUUACAAAAUAUAAUAAUACUCGUUUUUCGGUAGAGCGG---UAUUUUUAAGUCUAGGAUGUCAGAAAGGAUAUUCUAUAGAAAUACUCCAAAUUACAUCGUGACAU ..............((((................((((((.....))))))(---((((((((....(((((((((......)))))))))))))))))).............))))... ( -20.50) >DroSec_CAF1 57213 115 - 1 UGAUUUCGAUUAAAUUACAAAAUAUAAUAAUACUCGUUUUUCGGUAGAGCGG---UAUUCU--AGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGGCAUCAUGACAU ((((.(((......................(((.((((((.....)))))))---))((((--(...((((((((((....)))))))))))))))..........))).))))...... ( -25.80) >DroSim_CAF1 43708 115 - 1 UGAUUUUGAUAAAAUUACAAAAUAUAAUAAUACUCGUUUUUCGGUAGAGCGG---UAUUCU--AUUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCGAAAUGACAUCAUGACAU ..(((((((.....................(((.((((((.....)))))))---))((((--(...((((((((((....)))))))))))))))....)))))))............. ( -27.10) >DroEre_CAF1 56710 118 - 1 UGAUUUCAAUAAAAUUAAAAAGUAGUUUAUUACCAUAUUCUCAGUAAAGGGGGGCUAUUCU--AGUCUAGAAUGUCUAAAAGGACAUUCUAUAGGAAUACUCCAAAUGACAUCAUGACAU ((((.(((...((((((.....))))))......................((((.((((((--....((((((((((....))))))))))..))))))))))...))).))))...... ( -26.70) >consensus UGAUUUCGAUAAAAUUACAAAAUAUAAUAAUACUCGUUUUUCGGUAGAGCGG___UAUUCU__AGUCUAGAAUGUCUGAAAGGACAUUCUAUAGAAAUACUCCAAAUGACAUCAUGACAU ..(((((..........................(((((((.....)))))))...............((((((((((....))))))))))..)))))...................... (-16.55 = -16.05 + -0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:24 2006