
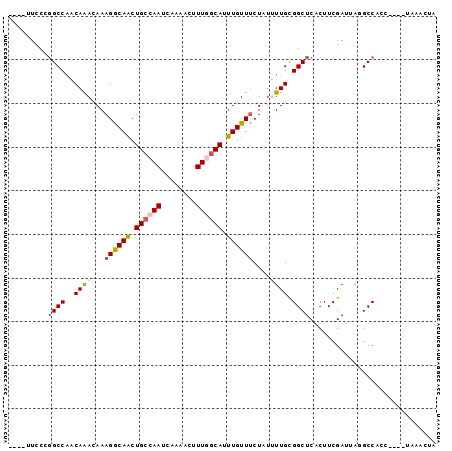
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,699,011 – 12,699,133 |
| Length | 122 |
| Max. P | 0.514823 |

| Location | 12,699,011 – 12,699,101 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -13.78 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12699011 90 - 22224390 ----UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUCCUAUUUUGCGGCUCACUUCGAUUAGGCCACC----UCAACUA ----.....((((..((((...((((((.((((((........)))))).)))..)))..))))(((......)))....))))...----....... ( -21.60) >DroPse_CAF1 65361 89 - 1 ----UUCCCGGCCACCAGACAAAGACAGCUGCCAAUCAAAAGUUUUGCAUUUGUUUCUAUGUUGCGGCUCACUUCGAU-ACGCCAUC----CAAGGCA ----.....((((....((((.((((((.(((.((........)).))).))))))...))))..)))).........-..(((...----...))). ( -18.30) >DroSec_CAF1 51844 90 - 1 ----UUCCUCGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAGUUGUUUCUAUUUUGCGGCUCUCUUCGAUUAGGCCACC----UCAACUA ----......(((..((((...(((((((((((((........)))))))))))))....))))(((......)))....)))....----....... ( -24.00) >DroEre_CAF1 51255 94 - 1 ----UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUUCUAUUUUGCGGCUCACUUCGAUUAGGCCACCUAUAUCAACUA ----.....((((..((((...((((((.((((((........)))))).))))))....))))(((......)))....)))).............. ( -22.20) >DroAna_CAF1 57629 92 - 1 UUCCUUUCCGGCCUACAAACAAAGGCAACUGCCAAUCAAAAGUUUGCCAUUUGUUUCUAUUUUGUGGCUCACUUCGAUUC-GCCACC-----AAACUA .........(((...........(((....)))((((..((((..(((((..((....))...)))))..)))).)))).-)))...-----...... ( -19.70) >DroPer_CAF1 80427 89 - 1 ----UUCCCGGCCACCAGACAAAGACAGCUGCCAAUCAAAAGUUUUGCAUUUGUUUCUAUGUUGCGGCUCACUUCGAU-ACGCCAUC----CAAGGCA ----.....((((....((((.((((((.(((.((........)).))).))))))...))))..)))).........-..(((...----...))). ( -18.30) >consensus ____UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUUCUAUUUUGCGGCUCACUUCGAUUAGGCCACC____UAAACUA .........((((..((((...((((((.((((((........)))))).))))))....)))).))))............................. (-13.78 = -14.28 + 0.50)


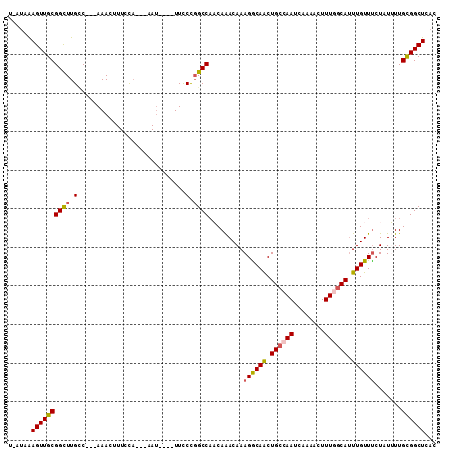
| Location | 12,699,033 – 12,699,133 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.95 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -17.83 |
| Energy contribution | -17.94 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12699033 100 - 22224390 U-AUAAAGUUGCGGCUUGCC---AAACUUUCUA---CAU----UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUCCUAUUUUGCGGCUCAC .-..((((((..((....))---.))))))...---...----.....((((..((((...((((((.((((((........)))))).)))..)))..)))).))))... ( -23.30) >DroPse_CAF1 65382 100 - 1 U-CUAAAGUUGCGGCUUGUC---AAAGUUUCCA---AAU----UUCCCGGCCACCAGACAAAGACAGCUGCCAAUCAAAAGUUUUGCAUUUGUUUCUAUGUUGCGGCUCAC .-....((((((((((((((---...((..((.---...----.....))..))..)))))((((((.(((.((........)).))).))))))....)))))))))... ( -21.00) >DroSec_CAF1 51866 100 - 1 U-AUAAAGUUGCGGCUUGCC---AAACUUUCUA---CAU----UUCCUCGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAGUUGUUUCUAUUUUGCGGCUCUC .-..((((((..((....))---.))))))...---...----......(((..((((...(((((((((((((........)))))))))))))....)))).))).... ( -29.40) >DroYak_CAF1 55429 100 - 1 U-AUAAAGUUGCGGCUUGGC---AAAGUUUCUA---CAU----UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUUCUAUUUUGCGGCUCAC .-....((((((((((.((.---(((((....)---).)----)))).)))).........((((((.((((((........)))))).)))))).......))))))... ( -28.10) >DroAna_CAF1 57649 111 - 1 CCCUGGAGUUGCGGUUUGCCGCAAAACUUUCCCAAAAACUUCCUUUCCGGCCUACAAACAAAGGCAACUGCCAAUCAAAAGUUUGCCAUUUGUUUCUAUUUUGUGGCUCAC ....((((((((((....))))))....))))................((((.((((((((((((((((..........)).))))).))))).......))))))))... ( -27.31) >DroPer_CAF1 80448 100 - 1 U-CUAAAGUUGCGGCUUGUC---AAAGUUUCCA---AAU----UUCCCGGCCACCAGACAAAGACAGCUGCCAAUCAAAAGUUUUGCAUUUGUUUCUAUGUUGCGGCUCAC .-....((((((((((((((---...((..((.---...----.....))..))..)))))((((((.(((.((........)).))).))))))....)))))))))... ( -21.00) >consensus U_AUAAAGUUGCGGCUUGCC___AAACUUUCCA___AAU____UUCCCGGCCAACAAACAAAGGCAACUGCCAAUCAAAACUUUGGCAUUUGUUUCUAUUUUGCGGCUCAC ......((((((((((.(............................).)))).........((((((.((((((........)))))).)))))).......))))))... (-17.83 = -17.94 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:20 2006