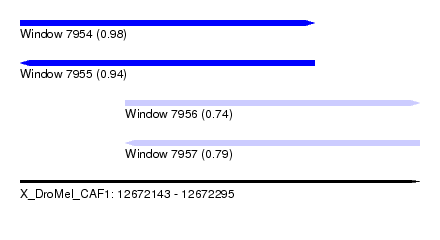
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,672,143 – 12,672,295 |
| Length | 152 |
| Max. P | 0.984251 |

| Location | 12,672,143 – 12,672,255 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 96.43 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -30.19 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12672143 112 + 22224390 CCCAAAGUGCUUUGGUGUGCGUAAAGGGCCAUUGUGAUUUUAAUUUUCAUGGCUCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCU .....((.(((((((((.((((...(((((((...(((....)))...)))))))((((((.....)))))).)))))))))))..(((((.((.....)))))))..)))) ( -31.90) >DroSec_CAF1 25765 110 + 1 GCCAAAGUGCUUUGGUGUGCGUAAAGGACCAUUGUGAUUUUAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGC-- ........(((((((((.((((...((.((((...(((....)))...)))).))((((((.....)))))).)))))))))))..(((((.((.....)))))))..))-- ( -26.40) >DroEre_CAF1 24976 112 + 1 UCCAAAGUGCUUUGGUGUGCGUGAAGGGCCAUUGUGAUUUUAAUUUUCAUGACCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCU .....((.(((((((((.((((((.(((...(..(((.........)))..)))).(((((.....))))))))))))))))))..(((((.((.....)))))))..)))) ( -31.50) >DroYak_CAF1 29116 112 + 1 CCCAAAGUGCUUUGGUGUGCGUGAAGGGCCAUUGUGAUUUUAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCU .....((.(((((((((.((((((.(((((((...(((....)))...))))))).(((((.....))))))))))))))))))..(((((.((.....)))))))..)))) ( -37.80) >consensus CCCAAAGUGCUUUGGUGUGCGUAAAGGGCCAUUGUGAUUUUAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCU ........(((((((((.((((((.(((((((...(((....)))...))))))).(((((.....))))))))))))))))))..(((((.((.....)))))))..)).. (-30.19 = -31.00 + 0.81)



| Location | 12,672,143 – 12,672,255 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.43 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12672143 112 - 22224390 AGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGAGCCAUGAAAAUUAAAAUCACAAUGGCCCUUUACGCACACCAAAGCACUUUGGG .((((.(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).)))).............))))...........(((((....))))). ( -29.80) >DroSec_CAF1 25765 110 - 1 --GCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUAAAAUCACAAUGGUCCUUUACGCACACCAAAGCACUUUGGC --..........(((..(((((.((((((((.(((((.((.((((((...)))))))).))))))))))))).........((((...........)))))))))..))).. ( -27.90) >DroEre_CAF1 24976 112 - 1 AGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGUCAUGAAAAUUAAAAUCACAAUGGCCCUUCACGCACACCAAAGCACUUUGGA .((((.(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).)))).............))))...........(((((....))))). ( -28.50) >DroYak_CAF1 29116 112 - 1 AGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUAAAAUCACAAUGGCCCUUCACGCACACCAAAGCACUUUGGG .((((.(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).)))).............))))...........(((((....))))). ( -31.40) >consensus AGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUAAAAUCACAAUGGCCCUUCACGCACACCAAAGCACUUUGGG .((((.(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).)))).............))))...........(((((....))))). (-28.75 = -28.62 + -0.12)


| Location | 12,672,183 – 12,672,295 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12672183 112 + 22224390 UAAUUUUCAUGGCUCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCUUGUAACCUAGUAUUAAAUGCACAUAAUUGAUUGUAUGCGG ...((((.(((((.(((((((.....))))).)).))))).)))).(((((((......(((.....))))))))))............((((.((((....)))).)))). ( -24.00) >DroSec_CAF1 25805 99 + 1 UAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGC-------------AUUAAAUGCACAUAAUUGAUUAUAGUCGG (((((((.(((((.(((((((.....))))).)).)))))..)))))))...(((...((((.....)))-------------).....))).......((((....)))). ( -21.20) >DroEre_CAF1 25016 112 + 1 UAAUUUUCAUGACCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCUUGUGCCCUAGUGUUCAACGCACAUUAUUGAUUAUAUUCGU ........((((...((((((((..........((((.........(((((.((.....))))))).......))))....((((.....)))).)))))))).....)))) ( -17.69) >DroYak_CAF1 29156 112 + 1 UAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCUCGUAACCUAGUAUUCAACGCACAUUAUUGAUUAUAUUCUU ...((((.(((((.(((((((.....))))).)).))))).)))).(((((.((.....)))))))...............(((.((((.........)))).)))...... ( -19.10) >consensus UAAUUUUCAUGGCCCGUCAAUAAAACAUUGAUCGCGCCAUCAAAAAUUACAAGCAAAUUGCUGUAAAAGCCUUGUAACCUAGUAUUAAACGCACAUAAUUGAUUAUAUUCGG ...((((.(((((.(((((((.....))))).)).))))).)))).(((((.((.....))))))).............................................. (-16.20 = -16.45 + 0.25)


| Location | 12,672,183 – 12,672,295 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12672183 112 - 22224390 CCGCAUACAAUCAAUUAUGUGCAUUUAAUACUAGGUUACAAGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGAGCCAUGAAAAUUA ..(((((.((....)).)))))............(((((((((((.....)))......))))))))((((.(((((.((.((((((...)))))))).))))).))))... ( -25.80) >DroSec_CAF1 25805 99 - 1 CCGACUAUAAUCAAUUAUGUGCAUUUAAU-------------GCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUA ............(((((((.(((.....(-------------(((.....))))...))).)))))))(((.(((((.((.((((((...)))))))).))))).))).... ( -24.10) >DroEre_CAF1 25016 112 - 1 ACGAAUAUAAUCAAUAAUGUGCGUUGAACACUAGGGCACAAGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGUCAUGAAAAUUA .................(((((.(((.....))).)))))......(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).))))... ( -27.00) >DroYak_CAF1 29156 112 - 1 AAGAAUAUAAUCAAUAAUGUGCGUUGAAUACUAGGUUACGAGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUA ....................((.((((((.....))).))).))..(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).))))... ( -26.00) >consensus ACGAAUAUAAUCAAUAAUGUGCAUUGAAUACUAGGUUACAAGGCUUUUACAGCAAUUUGCUUGUAAUUUUUGAUGGCGCGAUCAAUGUUUUAUUGACGGGCCAUGAAAAUUA ..............................................(((((((.....)).))))).((((.(((((.((.((((((...)))))))).))))).))))... (-20.91 = -20.73 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:05 2006