
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,665,815 – 12,665,913 |
| Length | 98 |
| Max. P | 0.701137 |

| Location | 12,665,815 – 12,665,913 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -19.67 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.66 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
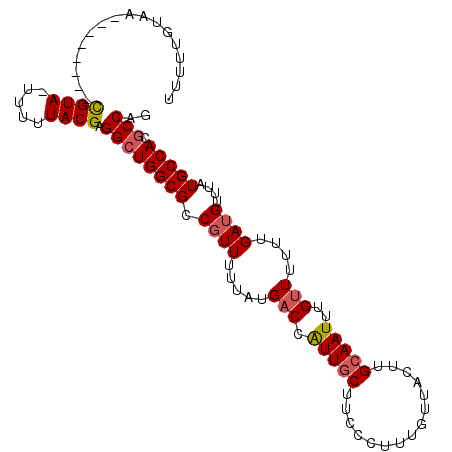
>X_DroMel_CAF1 12665815 98 + 22224390 UUUUUGUAA--------CGUAUUUUUUACGAGGCUGGCGCCGUUUUUAUGACCAUUGCUUCCCUUUGUUACUUGCAAUUUGUUUUUUGAUGUUUAUGCCACGCCAG .........--------((((.....)))).((((((((.((((.....(((.(((((...............)))))..)))....))))....))))).))).. ( -20.36) >DroSec_CAF1 19442 98 + 1 UUUUUGUAA--------CGUAUUUUUUACGAGGCUGGCGCCGUUUUUAUGACCAUUGCUUCCCUUUGUUACUUGCAAUUUGUUUUUUGAUGUUUAUGCCACGCCAG .........--------((((.....)))).((((((((.((((.....(((.(((((...............)))))..)))....))))....))))).))).. ( -20.36) >DroEre_CAF1 19675 93 + 1 ---UUUU-A--------CGUA-UUUUUACGAGGCUGGCGCCGUUUUUAUGACCAUUGCUUCCCUUUGUUACUUGCAAUUUGCUUUUUGAUGUUUAUGCCACGCCAG ---....-.--------((((-....)))).((((((((.((((.....(...(((((...............)))))...).....))))....))))).))).. ( -19.36) >DroAna_CAF1 24779 99 + 1 UUUUCGUAGUAGUAUUUUGUA-UUUUUACGAGGAUGGCGCC-UUUUUAUGACCGUUUCU----UUUGUUACUUGCAAUUUGUU-UUUGAUGUUUAUGCCACGCCAG (((((((((.(((((...)))-)).)))))))))(((((..-....(((((.((((.(.----.((((.....))))...)..-...)))).)))))...))))). ( -18.60) >consensus UUUUUGUAA________CGUA_UUUUUACGAGGCUGGCGCCGUUUUUAUGACCAUUGCUUCCCUUUGUUACUUGCAAUUUGUUUUUUGAUGUUUAUGCCACGCCAG .................((((.....)))).((((((((.((((.....(((.(((((...............)))))..)))....))))....))))).))).. (-14.03 = -14.66 + 0.63)


| Location | 12,665,815 – 12,665,913 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -16.45 |
| Consensus MFE | -12.44 |
| Energy contribution | -13.19 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12665815 98 - 22224390 CUGGCGUGGCAUAAACAUCAAAAAACAAAUUGCAAGUAACAAAGGGAAGCAAUGGUCAUAAAAACGGCGCCAGCCUCGUAAAAAAUACG--------UUACAAAAA ..(((.((((..................(((((...............))))).(((........)))))))))).((((.....))))--------......... ( -17.86) >DroSec_CAF1 19442 98 - 1 CUGGCGUGGCAUAAACAUCAAAAAACAAAUUGCAAGUAACAAAGGGAAGCAAUGGUCAUAAAAACGGCGCCAGCCUCGUAAAAAAUACG--------UUACAAAAA ..(((.((((..................(((((...............))))).(((........)))))))))).((((.....))))--------......... ( -17.86) >DroEre_CAF1 19675 93 - 1 CUGGCGUGGCAUAAACAUCAAAAAGCAAAUUGCAAGUAACAAAGGGAAGCAAUGGUCAUAAAAACGGCGCCAGCCUCGUAAAAA-UACG--------U-AAAA--- ..(((.((((..................(((((...............))))).(((........)))))))))).((((....-))))--------.-....--- ( -17.86) >DroAna_CAF1 24779 99 - 1 CUGGCGUGGCAUAAACAUCAAA-AACAAAUUGCAAGUAACAAA----AGAAACGGUCAUAAAAA-GGCGCCAUCCUCGUAAAAA-UACAAAAUACUACUACGAAAA .((((((.(((...........-.......)))..........----.((.....)).......-.))))))...(((((....-.............)))))... ( -12.20) >consensus CUGGCGUGGCAUAAACAUCAAAAAACAAAUUGCAAGUAACAAAGGGAAGCAAUGGUCAUAAAAACGGCGCCAGCCUCGUAAAAA_UACG________UUACAAAAA ..(((.((((..................(((((...............))))).(((........))))))))))..(((.....))).................. (-12.44 = -13.19 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:00 2006